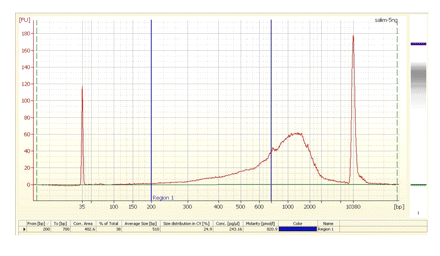
FIGURE 5.
Bioanalyzer profile of a typical nanoCAGE library (5 ng/μL). This protocol assumes that only cDNAs in the 200-700-bp range are sequenced efficiently. Longer templates are likely to be outcompeted by shorter ones for binding to the flow cell; they also could be difficult to amplify using the bridge PCR.










