Using Synthetic Peptide Substrates to Measure Drosophila Caspase Activity
Abstract
Central to the apoptotic pathway is the activation of caspases that are members of a highly conserved family of cysteine proteases. Caspases are synthesized as inactive zymogens and are generally activated by proteolytic cleavage to form the catalytically active enzyme. Caspase activity in apoptotic cells can be measured by assessing the cleavage of commercially available synthetic caspase substrates. The synthetic substrates contain a caspase cleavage site conjugated to a fluorochrome, such as 7-amino-4-methylcoumarin (AMC), or a chromophore, such as p-nitroaniline (pNA), for colorimetric detection. Here, we present a protocol for the measurement of caspase activity in Drosophila cell extracts by cleavage of the target peptide in the synthetic substrate that releases a fluorochrome or color-producing agent. The signal is measured by a spectrophotometer, with the intensity of the signal being proportional to the amount of substrate cleaved.
MATERIALS
Reagents
BCA Protein Assay Kit (Pierce)
Caspase substrate (e.g., VDVAD-AMC or DEVD-AMC fluorogenic substrates from MP Biomedicals)
Equipment
Benchtop centrifuge
Liquid nitrogen
Microcentrifuge tubes (1.5-mL)
Microtiter plates (96-well)
Pestle for 1.5-mL microcentrifuge tube
Spectrophotometer (e.g., FluoStar BD Biosciences)
METHOD
Preparation of Protein Lysates
-
1. Collect the samples (cell cultures, suitably staged whole animals, or dissected tissue) in 1.5-mL microcentrifuge tubes on ice in an appropriate volume of caspase assay lysis buffer.
-
The volume of lysis buffer can be adjusted depending on the protein yield of the sample and the abundance of active caspases—we use 10–20 µL per whole animal, 5 µL per dissected midgut tissue, and 20 µL to resuspend cells originally grown in a well of a six-well plate.
-
The samples can be snap-frozen in liquid nitrogen and stored at –70°C before the preparation of protein lysates.
-
-
2. Using a handheld pestle for a 1.5 mL microcentrifuge tube, homogenize the sample.
-
Approximately five strokes should be sufficient to disrupt cells/tissue and the sample will appear cloudy. If using whole animals, additional strokes may be required; the cuticle will remain and will be pelleted in the debris in Step 4.
-
-
3. Lyse the cells by freezing in liquid nitrogen and rapid thawing to room temperature three times.
-
4. Pellet the cell debris by spinning the extracts at 13,000 rpm (∼16,000g) in a benchtop centrifuge for 20 min at 4°C and then transfer the supernatant to a new tube.
-
5. Determine the protein concentration of the lysates.
-
We use the BCA Protein Assay Kit from Pierce (according to the manufacturer's instructions) as it is compatible with components of the caspase assay lysis buffer and works well with small amounts of lysate (dilute 1 µL in 50 µL water and use 10 µL for the assay).
-
If necessary, store the protein lysates at –70°C in aliquots.
-
Measuring Caspase Activity
-
The assays should be set up in triplicate and include the blank and calibration standards in a 96-well microtiter plate suitable for the spectrophotometer; a positive control, such as purified recombinant caspase, can also be included.
-
6. Set up the caspase assay on ice by combining 20–50 µg of protein lysate, caspase substrate VDVAD-AMC or DEVD-AMC at a concentration of 100 µm, and caspase assay lysis buffer for a final volume of 100 µL.
-
7. Prepare a blank control sample with substrate and no protein lysate.
-
8. Make serial dilutions of the substrate (5, 10, 25, 50, and 100 µm) in caspase assay lysis buffer.
-
These will be used to determine the fluorescence calibration standard.
-
-
9. Quantify the fluorescence for AMC using a spectrophotometer, with excitation at 385 nm and emission at 460 nm, over a time course (usually 3 h) at 37°C.
-
10. To obtain the conversion factor, plot the fluorescence calibration standards and obtain the slope of the standard curve.
-
The conversion factor is the inverse of the slope (1/slope of standard curve), and, for colorimetric substrates, is the standard concentration divided by the substrate blank reading.
-
-
11. Plot the change in fluorescence against time for each sample, adjusting for background for the sample by subtracting the zero time-point value from each time-point for each sample.
-
12. Determine the linear region of the slope of the line (m) from the formula
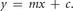
-
13. Calculate the caspase activity, expressed as pmol substrate/min, by multiplying the value of m by the conversion factor and then by the reaction volume (Denton et al. 2008; Kumar and Dorstyn 2009):
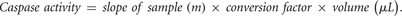
DISCUSSION
Caspase activity can vary greatly between different tissues and developmental stages, depending on the number of cells undergoing death at a given time. Caspases also play some nonapoptotic functions, so low levels of caspase activity can be present in many tissues. The amount of cell or tissue lysates to be used will need to be adjusted depending on these considerations. The protocol and reagents presented here are based on what we currently use in our laboratory. The assay is based on the measurement of active caspase proteolytic activity by cleavage of the target peptide in the synthetic substrate to release the fluorochrome or color-producing agent. There are many caspase peptide substrates available. We use Ac-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin (Ac-DEVD-AMC) and Ac-Val-Asp-Val-Ala-Asp-7-amino-4-methylcoumarin (Ac-VDVAD-AMC) as they show the highest level of activity for Drosophila caspases. Other newer and more-sensitive caspase substrates are available, but, as we have not used them, we have refrained from discussing them here.
ACKNOWLEDGMENTS
We thank the members of our laboratory for their comments on this protocol. The Drosophila cell death research in our laboratory is supported by the National Health and Medical Research Project Grant (1041807) and a Senior Principal Research Fellowship (1002863) to S.K.
Footnotes
-
↵1 Correspondence: Donna.Denton{at}unisa.edu.au; Sharad.Kumar{at}unisa.edu.au
- © 2015 Cold Spring Harbor Laboratory Press










