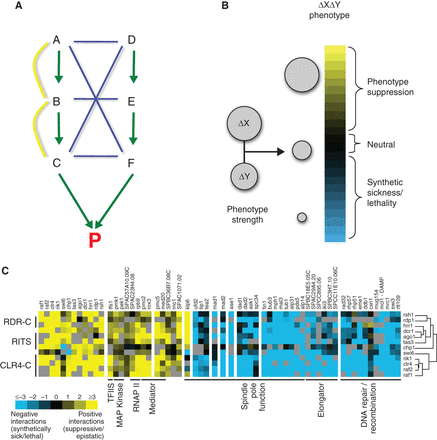
Epistasis mapping overview. (A) The logic behind synthetic sick/lethal (negative, blue edges) and buffering (positive, yellow edges) interactions. Two parallel pathways (A–B–C and D–E–F) lead to the same essential product (P). Different types of genetic interactions emerge, depending on how genes are disrupted in pairs. (B) Modeling genetic interactions, capturing the continuous spectrum of phenotype strengths. Single deletions (Δ of genes X and Y) are combined in a double-deletion strain. The expected fitness defect of the double mutant (ΔXΔY) is the product of the fitness defects of ΔX and ΔY. Genetic interactions are modeled as deviations from this expected value. The size of the circle is proportional to fitness. (C) Computational analysis of epistatic interactions. GI profiles of 12 members of the RNAi machinery in S. pombe are clustered. Positive interactions are observed within the pathway and with parts of the transcription machinery. Negative interactions are observed with genes involved in spindle function, DNA repair, and the elongator complex.










