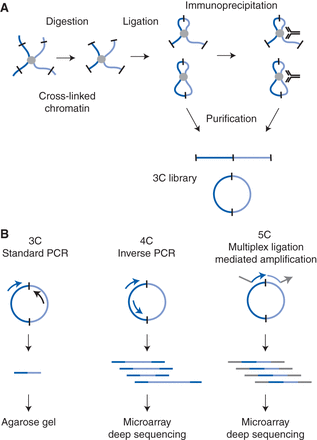
Schematic outline of 3C-based technologies. (A) Cross-linked chromatin segments (light blue and dark blue lines) connected by a protein complex (gray circle) are digested with a restriction enzyme (sites indicated by black bars) and then ligated. In the standard 3C protocol, DNA cross-links are then reversed and DNA is purified. In the ChIP-loop protocol, ligated chromatin is first immunoprecipitated with a specific antibody against a protein of interest, and then cross-links are reversed and DNA is purified. Both protocols yield a 3C ligation product library that is composed of both linear and circular DNA molecules. (B) Three methods for analysis of the 3C ligation product library. In the conventional 3C protocol, ligation products are detected by PCR with primers that amplify the ligation junction (primers are indicated with arrows; the arrowhead is the 3′ end of the primer). The amplified DNA fragments are detected and quantified on an agarose gel. In the 4C protocol, inverse PCR is used to amplify all fragments ligated to a single fragment of interest. The resulting amplified DNA is analyzed on a microarray or by deep (high-throughput) sequencing. In the 5C protocol, sets of ligation products are detected by multiplex ligation-mediated amplification. 5C primers are designed to anneal adjacent to each other across the ligation junction; arrowheads indicate the 3′ ends. Note the presence of universal tails (gray line/arrow) on the 5C primers that do not anneal to the 3C ligation product. See Figure 2 of details of primer design. The resulting amplified DNA is analyzed on a microarray or by deep sequencing.










