Analysis of Cell Fate Commitment in Xenopus Embryos
- Correspondence: samoody{at}gwu.edu
Abstract
The fates of individual cleavage-stage blastomeres and of groups of cells at the blastula through gastrula stages of Xenopus embryos have been mapped in great detail. These studies identified the major contributors of the three germ layers as well as a variety of tissues and organs and several specific cell types. One can use these fate maps to test the commitment of single cells or groups of cells to produce their normal repertoire of descendants, to identify the genes that regulate fate commitment, and to modulate the levels of gene expression in specific lineages to determine gene function in a variety of developmental processes. Here we introduce methods that include how to identify specific blastomeres, inject them with lineage tracers, and alter gene expression levels. We also discuss methods for assaying protein and mRNA expression in situ and for providing novel embryonic environments to test fate commitment. These techniques draw upon classical approaches that are quite easy to perform in the versatile Xenopus embryo.
FATE MAPPING AND LINEAGE TRACING
Fate mapping techniques have been used extensively in Xenopus to identify what kinds of tissues descend from a single blastomere or a specific region of the embryo. By precisely defining the regions of the embryo from which the germ layers, tissues and organs, and specific cell types arise, one can discover the mechanisms that control the organization of the body plan and specify cell fate. The creation of fate maps was recognized from the beginnings of experimental embryology as an essential tool (for review, see Klein and Moody [2016]). However, fate maps only describe the developmental path taken by a cell in the normal, intact embryo; they do not identify the full developmental potential of a cell or the mechanisms by which its fate is determined. Since cell fate can be influenced by a number of intrinsic and extrinsic factors, lineage tracing is an essential technique to identify the progeny of a cell as they develop under different experimental conditions. It also is an important tool for labeling donor tissue in tissue recombination experiments that are used to define inductive interactions. For example, the ability to recognize the descendants of the transplanted “organizer” in the host embryo was critical for interpreting the pioneering experiments of primary embryonic induction (Spemann and Mangold 1924).
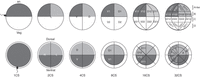
Lineage studies, fate mapping and tests of the fate determination of single blastomeres have been very informative in species in which the cell division patterns are identical across embryos. The invariant lineages allow the exact same cell to be marked in a large number of embryos (e.g., Sulston et al. 1983; Nishida 1997). In most vertebrate embryos, however, cleavage patterns are notably irregular (e.g., Kelly 1977; Ziomek et al. 1982; Johnson and Ziomek 1983; Kimmel and Law 1985). In contrast, Xenopus embryos often cleave in nearly identical patterns, and a few common cleavage patterns have been described (Fig. 1; Hirose and Jacobson 1979). Another essential element for accurate fate mapping is the ability to identify the body axes (dorsal–ventral, left–right, anterior–posterior) at early cleavage stages so that a blastomere's position can be ascertained. Due to unique movements of pigment granules after fertilization, this is possible in Xenopus. The first cleavage furrow, which always defines the mid-sagittal plane that separates the right and left sides, naturally bisects the lightly pigmented gray crescent region of the animal hemisphere in about 70% of embryos. By picking these embryos at the two-cell stage, one can use the dorsal–ventral differences in pigmentation to identify specific blastomeres at later stages (Klein 1987; Masho 1990). By choosing embryos from this subset that also cleave in a “stereotypic” pattern, one can study nearly identical lineages across a number of embryos (see Protocol: Lineage Tracing and Fate Mapping in Xenopus Embryos [Moody 2018a]). This is of significant experimental advantage because it decreases variability between embryos and enables the construction of robust qualitative and quantitative fate maps.
Diagram indicating the pigmentation differences and nomenclature of cells from the one-cell to the 32-cell stage. The top row represents a left (L) side view. The bottom row represents an animal pole view. The point of sperm entry is indicated at the one-cell stage. an, animal pole; veg, vegetal pole; CS, cell stage; D, dorsal; V, ventral; L, left; R, right. Nomenclature is that of Jacobson (Hirose and Jacobson 1979; Jacobson and Hirose 1981). Tiers on the 32-cell embryos refer to the Nakamura and Kishiyama (1971) nomenclature.
Detailed fate maps of the different cleavage-stage blastomeres of Xenopus using vital dyes and intracellular lineage tracers have been published (Nakamura and Kishiyama 1971; Hirose and Jacobson 1979; Jacobson and Hirose 1981; Dale and Slack 1987; Moody 1987a,b, 1989; Masho 1988; Masho and Kubota 1988; Moody and Kline 1990). They show that when the same, stereotypic blastomere is traced in a large number of embryos, its fate is quite predictable, in contrast to most other vertebrates (e.g., Kelly 1977; Ziomek et al. 1982; Johnson and Ziomek 1983; Kimmel and Law 1985). This predictability allows one to rigorously test how and when during development a particular fate is determined.
The Xenopus blastomere fate maps demonstrate some important features of fate restriction during cleavage stages. First, there is no germ layer restriction at the two- through 32-cell stages; all blastomeres give rise to some cells that reside in the endoderm, mesoderm, and ectoderm. However, as expected from the gastrula fates maps (Keller 1975, 1976), vegetal blastomeres contribute more prominently to endoderm, equatorial blastomeres more prominently to mesoderm, and animal blastomeres more prominently to ectoderm. A second important feature is that no tissue or organ has been found to descend from a single blastomere, i.e., none is monogenic. However, when one places the fate map data from two-cell to 32-cell embryos in a true lineage map, i.e., a map of mitotic relationships within the clone, some apparent restrictions are revealed (Moody and Kline 1990). For example, within the ectoderm, the cement gland, lens, olfactory placode, cranial ganglia, and otocyst are derived from animal (D1, V1) but not from vegetal (D2, V2) eight-cell blastomeres. Rostral CNS structures (retina, forebrain, and midbrain) are restricted to the D cell at the four-cell stage, and further restricted to its anterior daughter (D1) at the four-cell stage. But at the following two cell divisions (the 16-cell and 32-cell stages), all daughters and granddaughters contribute to all three structures. Thus, germ layer, tissue, and organ fates are not restricted to a single cleavage-stage blastomere, at least through the 32-cell stage. Nonetheless, there are regional differences that allow one to target manipulations to major organs or regions. For example, at the 16-cell stage one can target different ectodermal structures. The major contributor to the retina is blastomere D1.1 and the major contributor to the epidermis is blastomere V1.1. The neural crest derives mostly from blastomere D1.2 and the cranial placodes mostly from blastomere V1.2 (Fig. 1; Moody 1987a).
One also can test cell fate restriction at the cell phenotype level by labeling cells with cell type–specific markers (see Protocol: Whole-Mount Immunocytochemistry in Xenopus [Klymkowsky 2018]). However, even with these very specific approaches we have found no evidence for a monoclonal origin of several neuronal phenotypes. For example, primary motor neurons and primary sensory neurons, which can be identified by their very characteristic morphologies when filled with a cytoplasmic lineage tracer (Moody and Jacobson 1983; Jacobson and Moody 1984), each descend from a large number of 32-cell blastomeres (Moody 1989). We thought that this polyclonal origin might be related to their extensive anterior–posterior distribution along the length of the spinal cord, so we next studied several neuronal phenotypes that are spatially restricted. However, these cells also descend from multiple blastomeres. For example, four different neurotransmitter types of retinal amacrine cells each descend from several 32-cell blastomeres (Huang and Moody 1995, 1997). Even a small dopaminergic nucleus in the hypothalamus, which contains only 30–40 cells at early tadpole stages, descends from as many as seven of the 32 blastomeres (Huang and Moody 1992). Further studies using blastomere transplantation and deletion approaches (see Protocol: Cleavage Blastomere Deletion and Transplantation to Test Cell Fate Commitment in Xenopus [Moody 2018b]) showed that the ability of a blastomere to contribute to the retina depends on both intrinsic factors and environmental cues (Yan and Moody 2007). These studies illustrate the power of combining single cell lineage labeling with cell phenotype markers to explain how cell fate is specified.
TESTING FATE COMMITMENT
Fate maps, however, do not describe the full developmental potential of a blastomere or indicate whether it will express its specific cell fate regardless of the surrounding cellular environment. A cell that is committed to its fate will produce its normal repertoire of descendants even when it develops in other regions of the embryo. Alternatively, a cell that is not yet committed will express different cell fates depending upon the environment that it occupies. There are several ways to test the commitment of a blastomere to produce its mapped fate. One approach is to delete a neighboring blastomere to test if its presence is necessary for the normal fate of the cell of interest, and to determine whether the deleted lineage can be reconstituted (e.g., Huang and Moody 1993; Gallagher and Moody 2004) (see Protocol: Cleavage Blastomere Deletion and Transplantation to Test Cell Fate Commitment in Xenopus [Moody 2018b]). Another approach is to transplant a labeled blastomere or group of cells (e.g., Spemann and Mangold 1924; Yan and Moody 2007) into a novel region of the embryo to test whether it maintains its original set of descendants, or alters its progeny in accord with its new environment. Explanting a single blastomere or a group of cells (e.g., the animal cap) out of the embryo and into culture prevents the cell from communicating with its normal neighbors and allows one to experimentally manipulate extrinsic signals (see Protocol: Cleavage Blastomere Explant Culture in Xenopus [Moody 2018c] and Protocol: Dissecting and Culturing Animal Cap Explants [Dingwell and Smith 2018]). For each approach, one can analyze gene expression and the production of specific cell types using standard techniques of lineage tracing and in situ detection of cell type–specific mRNAs and proteins (see Protocol: Whole-Mount In Situ Hybridization of Xenopus Embryos [Saint-Jeannet 2017] and Protocol: Whole-Mount Immunocytochemistry in Xenopus [Klymkowsky 2018]).
ALTERING GENE EXPRESSION
Fate maps also do not identify the genes that regulate the development of a lineage. To do this, one needs to modify the gene expression of the precursor cell of interest. In Xenopus, this can be easily accomplished by microinjecting molecules into an identified blastomere of known fate to affect gene expression specifically within its lineage. One can microinject a number of different kinds of constructs, including mRNAs, plasmid DNAs, and antisense oligonucleotides (see Protocol: Microinjection of Xenopus Embryos with mRNAs and Oligonucleotides [Moody 2018d]) and Protocol: Microinjection of DNA Constructs for Gene Mis-Expression and Cis-Regulatory Module Analysis [Yasuoka and Taira 2018]). With these reagents one can ascertain whether a gene promotes or represses target genes or specific tissue formation, or converts tissues to novel fates when ectopically expressed. One can identify functional domains by expressing mutated forms and determine the requirement of a gene in a specific tissue or process by injecting dominant-negative forms or antisense oligonucleotides that block either the mRNA splicing or translation of endogenous proteins. For all of these constructs, it is essential to mark the blastomere that is injected with a lineage label to ascertain whether the effect is confined to the descendants of the injected blastomere (i.e., is cell autonomous) or involves surrounding cells (i.e., involves cell-to-cell interactions).
CONCLUSION
Together, the methods described here provide simple yet powerful ways to demonstrate the fundamental developmental mechanisms that regulate how the different cells and regions of the embryo produce specific cell types, tissues, and organs.