Genetic Toolbox Approaches in Mosquitoes
- ↵2Correspondence: olena.riabinina{at}durham.ac.uk
Abstract
Mosquitoes are fascinating and diverse study organisms that are rapidly becoming genetically tractable. Here, we review the latest and most commonly used approaches to silence or edit genes and express transgenes in mosquitoes. We also focus on gene drives—a genetic technology that may prove essential for combating mosquito-borne diseases.
INTRODUCTION
Mosquitoes are not genetic model organisms. However, they attracted intense research interest because of their ability to transmit zoonotic diseases, and the first transgenic mosquitoes were created more than 20 years ago (Coates et al. 1998; Jasinskiene et al. 1998; Catteruccia et al. 2000; Grossman et al. 2001; Perera et al. 2002). Since then, the genome editing field has been revolutionized by CRISPR–Cas9, which was quickly and successfully implemented in eight mosquito species (Gantz et al. 2015; Kistler et al. 2015; Dong et al. 2018; Li et al. 2018; Anderson et al. 2019; Duvall et al. 2019; Liu et al. 2019; Raji et al. 2019; Greppi et al. 2020; Li et al. 2020a, 2021; Meuti and Harrell 2020; Redhai et al. 2020; Sun et al. 2020; Feng et al. 2021a,b; Inbar et al. 2021; Purusothaman et al. 2021; Quinn et al. 2021; Zhan et al. 2021). It has also become apparent that although mosquito genetic tools are often instructed by similar tools in Drosophila, the Drosophila solutions cannot be ported directly into mosquitoes and need to be adjusted for each mosquito species of interest. Efforts to eliminate mosquito species that transmit malaria, dengue, and West Nile virus (Anopheles spp., Aedes spp., and Culex spp., respectively) have fueled the development of gene drives in mosquitoes more than in any other organism to date (Gantz et al. 2015; Hammond et al. 2016, 2017, 2021a; Kyrou et al. 2018; Adolfi et al. 2020; Li et al. 2020b; Hoermann et al. 2021). Here, we review recent progress in gene silencing by RNAi, genome editing and transgenesis, and the development of CRISPR–Cas9 gene drives. We have highlighted the most widely used or promising approaches that may be of use to mosquito researchers, focusing on the genetic techniques rather than on the original research questions.
GENE SILENCING BY RNAi
The suppression of gene expression can be induced by RNA interference (RNAi), as first observed in the nematode worm Caenorhabditis elegans (Fire et al. 1998). RNAi has now been used in Aedes aegypti (Hapairai et al. 2017), Aedes albopictus (Gu et al. 2011), Culex pipiens (Lopez-Martinez et al. 2012), Anopheles gambiae (Taracena et al. 2019), and Anopheles stephensi (Brown et al. 2003) mosquitoes, and may be implemented via delivery of double-stranded RNA (dsRNA), small interfering RNA (siRNA), or short hairpin RNA (shRNA). The RNAi process can be broken into three critical stages. First, a long dsRNA molecule that is introduced into the cell is cut into smaller RNA fragments by a Dicer or Ribonuclease III enzyme (RNase III). These smaller fragments are known as siRNAs and have a complementary sequence to the target mRNA (Zhang et al. 2004). Second, the siRNAs complex with the RNA-induced silencing complex (RISC), and the sense strand of the siRNA is degraded, leaving the antisense strand complexed (Kim and Rossi 2008). Third, the RISC-antisense siRNA strand complex binds to the complimentary target mRNA sequence before translation can occur and degrades the mRNA, preventing the expression of the gene (Fire et al. 1998). There are three primary methods of delivering RNAi into mosquito cells: (1) microinjection (e.g., Liu et al. 2010; Singh et al. 2013); (2) feeding (e.g., Zhang et al. 2010; Mysore et al. 2017); and (3) microbial delivery (Taracena et al. 2019).
Microinjection
Microinjection has been the most common method to induce RNAi in mosquitoes, particularly at the pupal and adult stages of development (Regna et al. 2016; Isoe et al. 2019). The advantage of using microinjection is that it ensures the delivery of dsRNA directly into the tissue of choice, avoiding the cuticle, gut epithelium, and hardy outer exoskeleton. Another benefit of microinjections is knowing precisely how much dsRNA is delivered into the organism, unlike feeding, in which the amount of dsRNA ingested may be unknown (Yu et al. 2013). However, the disadvantage of using microinjections is that they require a high degree of skill and may cause injury, thus affecting survival and experimental readouts. For example, Blitzer et al. (2005) targeted sterol carrier protein-2 (AeSCP-2) expression by injecting chemically synthesized and HPLC-purified dsRNA into adult and fourth-instar larvae of Ae. aegypti. Knockdown of AeSCP-2 in fourth instars reduced cholesterol accumulated in pupae by 33% but caused 42% adult mortality in the first 3 d of emergence.
Microinjection is rarely used in the larvae because of the small size of the larvae and increased likelihood of fatal injury to the organism. However, Liu et al. (2010) used this approach to down-regulate An. gambiae olfactory receptors 7 (AgOr7) and 40 (AgOr40), and An. gambiae variant ionotropic receptor 76b (AgIr76b) in fourth-instar larvae. Quantitative analysis showed a significant transcript level reduction of AgOr7, AgOr40, and AgIr76b after siRNA treatment. Larvae injected with the siRNA had a significantly reduced behavioral response to DEET compared to the control.
Whyard et al. (2015) injected Ae. aegypti pupae with dsRNA to target 10 genes that regulate fertility in adult males (see Supplemental Table S1). A quantitative reverse transcription polymerase chain reaction (qRT-PCR) confirmed that each of the genes was down-regulated by 70%–95% in the adult males 4 d postinjection and in total, nine out of 10 of the dsRNAs induced sterility in half of the males. Similarly, Meuti et al. (2015) microinjected adult female C. pipiens with dsRNA to knock down several circadian clock genes (per, tim, cry2, cyc, and pdf) to measure the effect on egg follicle length and lipid content. Reduction of mRNA expression by RNAi had varied success per gene: per (40%), tim (25%), cry2 (45%), cyc (15%), and pdf (60%).
It appears that the effects of RNAi can be highly variable not only between studies, but also between targets of the same study in the same species of mosquito. The duration of RNAi effects is also unclear. We thus suggest that down-regulation of selected genes should be experimentally validated for each target gene. In summary, delivery of dsRNA by microinjection is a suitable method that has been successfully used in, for example, An. gambiae (Michel et al. 2006; Liu et al. 2010; Liu and Zwiebel 2013; Regna et al. 2016), An. stephensi (Brown et al. 2003), Ae. aegypti (Blitzer et al. 2005; Berois et al. 2012; Erdelyan et al. 2012; Hapairai et al. 2017; Isoe et al. 2019), and Cx. pipiens (see Supplemental Table S1; Meuti et al. 2015).
Feeding or Soaking in a ds/siRNA Solution
Feeding or soaking is the most straightforward delivery method because of its simplicity and the lack of injury caused to the insect. This method is well-established, quick, repeatable, and convenient for larval and pupal stages with effects that can persist into the adult stage. The disadvantage of soaking is that large volumes of dsRNA needed to make it effective. Moreover, a study by Fischer et al. (2017) showed that dsRNA quickly degrades in aquatic environments, giving it limited application to larviciding in the field.
Singh et al. (2013) were the first to perform soaking on mosquito larva. They successfully targeted β-tubulin, chitin synthase-1 and -2, and heat shock protein 83 by soaking first-instar larvae of Ae. aegypti in H2O containing β-tub-dsRNA for 2 h before returning them to fresh H2O. The authors confirmed 100% uptake of the dsRNA solution by adding a blue dye that could be seen in the gut of the larvae when ingested. The length of time to soak a sample varies between developmental stages. Figueira-Mansur et al. (2013) soaked second-instar Ae. aegypti larvae for 24 h to silence the insecticide resistant AaegP-gp gene. Larvae soaked in the dsRNA solution had a 30% higher mortality in insecticides than control groups (see Supplemental Table S1).
Inducing RNAi in larvae may have effects that persist into the adult stage. Whyard et al. (2015) soaked larvae in a dsRNA solution for 1 h over 6 d to successfully target genes that regulate male fertility (bol, fzo, gas8, nht, and zpg). They used dsRNA at two concentrations: 0.02 mg/mL (which rendered 20%–35% of males sterile) and 0.2 mg/mL (which rendered 72%–92% of males sterile). The same method was used by the authors to target the female isoform of the sex determination gene doublesex, creating a male-biased population. Similarly, Lopez-Martinez et al. (2012) soaked dehydrated fourth-instar C. pipiens larvae in H2O containing dsRNA to down-regulate heat shock protein 90 (hsp90). The effects persisted through to the pupal and adult stages, with hsp90 levels reduced by 77% on the third day of adult life.
There have been many studies aimed at optimizing ds/siRNA delivery by feeding to adult mosquitoes. Mysore et al. (2020) targeted the gene Shaker, which when knocked down, produced lethal phenotypes and neural defects that affected coordination and prevented flight behavior in dsRNA-treated mosquitoes. RNAi targeting the dop1 gene has now been trialed as a pesticide delivered to adults in attractive toxic sugar baits (Hapairai et al. 2020). The study found that 88% of mosquitoes that fed on the sugar bait died and the remaining 12% could not fly. Toxic sugar bait studies thus show promise for the control of mosquito populations in the field (see Supplemental Table S1).
Chitosan Nanoparticle and Agarose Gel Delivery
The instability of dsRNA after delivery is a major difficulty for RNAi applications. The degradation of dsRNA may be delayed by encapsulating the ds/siRNA in chitosan nanoparticles (Howard et al. 2006) that may then be fed to mosquitoes. Mysore et al. (2013) used chitosan/siRNA nanoparticle feeding to investigate the role of the gene semaphorin-1a (sema1a) in the olfactory development of Ae. aegypti. Antennal lobe defects were detected in 44% of treated larvae and 32% of pupae. In the behavioral assay, 48% of treated larvae failed to touch the yeast pellet, a normally attractive food stimulus for the larvae. In another study, researchers disrupted cuticle formation in larvae and induced mortality in adult Ae. aegypti by feeding the mosquitoes chitosan/dsRNA nanoparticles to target the gene 3, 4-dihydroxyphenylacetaldehyde (DOPAL) synthase (Chen et al. 2019). Furthermore, Ramesh et al. (2016) compared two methods of dsRNA delivery to the third-instar larvae of Ae. aegypti. First, they fed the larvae bacterially expressed dsRNA, and second, they fed them chitosan/dsRNA nanoparticles to target the vestigial gene (vg).There was no significant difference in the efficiency of dsRNA delivery by either method, and both techniques resulted in high adult deaths and wing abnormalities.
Chitosan nanoparticles may also be used in combination with agarose gel to deliver RNAi through feeding. Zhang et al. (2010) reduced the transcript levels of two chitin synthase genes, AgCHS1 (by 62.8%) and AGCH2 (by 33.8%), by feeding third-instar larvae of Ae. aegypti with dsRNA coated in a mixture of agarose gel, which was further entrapped in chitosan/AgCHS dsRNA-based nanoparticles (see Supplemental Table S1). Incorporating RNAi into chitosan nanoparticles coated with an agarose gel helps protect the dsRNA from breaking down in the environment, as previously shown in mice (Gao et al. 2009). The technique is particularly well-suited for larval feeding because larvae eat almost continuously and because without the gel/particle protection, RNA quickly degrades in H2O.
Microbial Delivery (Yeast, Bacteria, Viruses, and Algae)
RNAi delivery through microbes has been developed in recent years as another method of oral delivery. Mysore et al. (2017) used engineered Saccharomyces cerevisiae that produced shRNAs to silence suppressor of actin (Sac1), leukocyte receptor complex member (Irc), and offtrack (Otk) by feeding the yeast to An. gambiae larvae. The knockdown of these genes resulted in a loss of neuronal synapses and 100% of larval mortality in An. gambiae.
Bacteria have also been used as an effective method of delivering RNAi to mosquito. Taracena et al. (2019) knocked down expression of the female-specific isoform of the doublesex gene AgdsxF in An. gambiae larvae through oral delivery of bacteria that were producing dsRNA. Feeding AgdsxF dsRNA reduced the mRNA transcript by 66% in females, and only half of the females survived to adulthood. Similar to the protection that chitosan nanoparticles give to dsRNA, microbial delivery offers RNAi protection against RNases and from breaking down in the gut of insects (Wiltshire and Duman-Scheel 2020).
Viral delivery has been used as another delivery method through soaking. Gu et al. (2011) developed a recombinant Ae. aegypti Densovirus (AeDNV) siRNA expression system that was introduced to Ae. albopictus by soaking second-instar larvae in H2O containing the virus stock. siRNA entered the body through the anal papillae before spreading to the muscle fibers, midgut, salivary glands, and several other tissues, down-regulating V-ATPase expression by up to 70% (see Supplemental Table S1).
Microalgae are one of the primary sources of nutrition for larvae in aquatic environments, therefore using transgenic algae as a vector for delivering dsRNA to the larvae has offered some potential. The advantage of microalgae is that they can be heat-killed, reducing the amount of genetically modified organisms released in the wild. Kumar et al. (2013) fed first-instar larvae of An. stephensi transgenic microalgae, targeting 3-hydroxykynurenine transaminase (3-HKT), a gene that catalyzes the transamination of 3-hydroxykynurenine (3-HK) to xanthurenic acid in the tryptophan catabolism pathway, resulting in an increase in reactive oxygen species that can kill the mosquito. The study found that dsRNA-treated larvae had 40% greater mortality than the control in every bioassay. Mortality was first observed in the second-instar stage, whereas treated larvae had significantly reduced levels of 3-HKT transcripts.
In summary, microbial delivery shows great potential because of its ease of manipulation, application to larviciding in the field, and ability to protect dsRNA from degradation in aquatic environments.
Transgenic Delivery
The use of transgenes to express dsRNA is a commonly used approach in Drosophila melanogaster, and a large collection of commercially available RNAi lines is currently available (Dietzl et al. 2007). Transgenic delivery has been successfully used in An. gambiae to down-regulate cyp4g16 and cyp4g17 genes involved in the production of cuticular hydrocarbons (Lynd et al. 2019). In their study, Lynd and colleagues (2019) used an oenocyte-specific driver to express cyp4g16/17 dsRNA, leading to mortality or high susceptibility of mosquitoes to desiccation during the pupal stage. The advantage of this transgenic approach is the ease of delivery via a genetic cross, which is suitable for research applications. However, establishing an efficient RNAi line may be tricky, and breeding several test lines is labor-intensive. Nevertheless, efficient RNAi lines could potentially be applied in the field by using gene drives (see the section CRISPR–Cas9 Gene Drives) for transgene propagation.
GENE EDITING
Gene editing in mosquitoes has so far been used to primarily study mosquito olfaction and the mechanisms of disease transmission (for reviews, see Dong et al. 2021; Konopka et al. 2021; Wheelwright et al. 2021), as well as for population disruption or replacement (see, e.g., Adolfi and Lycett 2018; Raban et al. 2020 and the section CRISPR–Cas9 Gene Drives). All these studies were published after 2012, when CRISPR–Cas9 was introduced as a tool for gene editing (Jinek et al. 2012). Therefore, it is not surprising that only a few studies used early gene editing tools, such as transcription activator–like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs). Notable examples of pre-CRISPR studies are works on chemosensation and thermosensation in Ae. aegypti that used ZFNs to mutate olfactory receptor coreceptor AeORCO (Degennaro et al. 2013), a CO2 coreceptor gene AeGR3 (McMeniman et al. 2014), and a temperature receptor TRPA1 (Corfas et al. 2015). The use of TALENs in mosquitoes was pioneered in Ae. aegypti by a proof-of-principle study that mutated the kmo gene, a gene essential for eye pigmentation (Aryan et al. 2013). At almost the same time, TALENs were used to mutate an immunity gene TEP1 in An. gambiae (Smidler et al. 2013). Although TALENs and ZFNs are still an option for creating DNA breaks at a predefined location, the low cost and ease of use of CRISPR–Cas9 has made it the gene editing tool of choice for mosquito researchers.
To date, CRISPR–Cas9 has been used in eight mosquito species: An. gambiae (Dong et al. 2018; Greppi et al. 2020; Redhai et al. 2020; Sun et al. 2020), An. stephensi (Gantz et al. 2015; Inbar et al. 2021), Anopheles funestus (Li et al. 2018; Quinn et al. 2021), Anopheles albimanus (Li et al. 2018), Ae. aegypti (Kistler et al. 2015; Duvall et al. 2019; Raji et al. 2019; Zhan et al. 2021), Ae. albopictus (Liu et al. 2019), Culex quinquefasciatus (Anderson et al. 2019; Li et al. 2020a; Feng et al. 2021a,b; Purusothaman et al. 2021), and C. pipiens (Meuti and Harrell 2020; Li et al. 2021), all of which are vectors of human disease (see Supplemental Table S2). CRISPR–Cas9 was used in these species to create null mutants via nonhomologous end joining (NHEJ) repair of Cas9-induced DNA breaks, and/or to introduce transgenes via homology-directed repair (HDR) of DNA breaks. Kistler et al. (2015) provide an excellent step-by-step method for CRISPR–Cas9 gene editing in Ae. aegypti, which, briefly, consists of injecting mosquito embryos with (1) Cas9 protein or mRNA, (2) sgRNA, and (3) ssDNA or dsDNA plasmid donor for HDR. Injections of recombinant Cas9 protein led to the highest rate of mutagenesis, up to 30%, followed by Cas9 mRNA. The authors also tried injecting a plasmid containing a polyubiquitin_promoter-Cas9 construct but found it ineffective. The rate of HDRs observed was also up to 30%. A somewhat different method was used for Anopheles (Hammond et al. 2016; Sun et al. 2020; Quinn et al. 2021) and Ae. aegypti (Zhu et al. 2021). The authors used Vasa promoter, which is active in the germline, to drive expression of Cas9 at the early embryonic stage. Vasa_promoter-Cas9 and U6_promoter-sgRNA constructs were cloned into the same plasmid that was injected into the mosquito embryos together with a second plasmid containing a homology repair template. This technique led to a 2%–7% rate of HDR insertions in Anopheles, indicating that the Vasa promoter can efficiently drive Cas9 expression in the germline.
Chaverra-Rodriguez et al. (2018) have pioneered ReMOT Control (Receptor-Mediated Ovary Transduction of Cargo)—an alternative, high-throughput method of CRISPR component delivery based on the phenomenon known as maternal deposition, whereby protein material is synthesized by the mother and delivered to the embryos. In the ReMOT Control method, a carrier peptide P2C is fused to the Cas9 protein and injected into the abdomen of a gravid female together with sgRNA and an endosomal escape reagent. The P2C peptide ensures that the Cas9 + sgRNA complex is taken up by the embryo from the mother's hemolymph, and the endosomal escape reagent ensures the Cas9 + sgRNA complex is released from the uptake endosome and reaches the embryo's nuclei. This method bypasses one of the main bottlenecks of mosquito genetic modification: the labor-intensive manual injection of individual embryos (Bui et al. 2020; Meuti and Harrell 2020; Carballar-Lejarazú et al. 2021). ReMOT Control has been successfully used in Ae. aegypti (Chaverra-Rodriguez et al. 2018), An. stephensi (Macias et al. 2020), and C. pipiens (Li et al. 2021) to produce knockout mutants with success rates as high as 0.3 mutants per injected female. No knock-ins or random transgene insertions were yet reported with ReMOT Control, but when further developed, this method will make in-house mosquito genetic editing and transgenesis available to numerous laboratories.
TRANSGENES AND BINARY EXPRESSION SYSTEMS
Transgenes
The ability to introduce transgenes, instead of only creating mutations, takes genetic editing of mosquitoes to the next level. Interestingly, random insertions of transgenes were successfully achieved in An. stephensi (Catteruccia et al. 2000), An. gambiae (Grossman et al. 2001), An. albimanus (Perera et al. 2002), and Ae. aegypti (Coates et al. 1998; Jasinskiene et al. 1998) decades before successful gene editing (see Supplemental Table S2). Random insertions of transgenes into Aedes fluviatilis (Rodrigues et al. 2006) and Ae. albopictus (Labbé et al. 2010) followed soon after. More recently, piggyBac and Mos vectors were successfully used for random transgene insertions in An. gambiae (Riabinina et al. 2016) and Ae. aegypti (Buchman et al. 2019; Bui et al. 2019; Aryan et al. 2020; Shankar et al. 2020). Transgenes may also be integrated into the “docking sites” of known genomic locations, such as pattB (Franz et al. 2011; Meredith et al. 2011, 2013). The pattB sites are first integrated as random insertions into the genome, and then their position is mapped out and characterized for strength of expression and leakiness. The pattB insertion may be preferred to a random insertion when researchers do not need to obtain a number of transgenic lines with different strengths of transgene expression. The transgene often consists of a mosquito-active promoter fused to the reporter of interest, such as green fluorescent protein (GFP). The transgenes also usually carry an easily visible marker for screening. In contrast to Drosophila, in which curly wings, number and shape of bristles, eye color and shape, etc., serve as markers, mosquito markers are limited and consist of a fluorescent protein driven by an eye (3xP3) or a midgut (actin5C) enhancer and promoter (Volohonsky et al. 2015; Bernardini et al. 2018; Webster and Scott 2021). Thus, having good, easily visible markers in mosquitoes is important—genetic crosses of several transgenic or mutant lines would be greatly simplified if each had its own visual marker. Volohonsky et al. (2015) provide a useful compilation of tools that they developed for Anopheles transgenesis, including expression patterns of commonly used markers and reporters, and An. gambiae codon usage preference.
Binary Expression Systems
Binary systems for transgene expression have proved to be effective and popular in mosquitoes (Driesschaert et al. 2021 offers an excellent recent introduction to transgene expression and overview of binary expression systems). Briefly, binary expression systems have two key components: One determines in which cells and at what times a transgene is expressed; the other carries the coding sequence of the transgene to be expressed. Binary systems provide several advantages over promoter-reporter fusions: (1) The expression of the reporter gene will be greatly amplified; (2) different reporters/effectors will always be expressed in the same pattern if the same driver is used; and (3) fewer transgenic lines need to be maintained as soon as the number of promoters and reporters exceeds two of each. So far, three binary systems have been introduced into mosquitoes: TetOn-Off (Gossen and Bujard 1992; Lycett et al. 2004), GAL4/UAS (Brand and Perrimon 1993; Kokoza and Raikhel 2011), and the Q-system (Potter et al. 2010; Riabinina et al. 2015, 2016).
The GAL4/UAS system was piloted in Ae. aegypti (Kokoza and Raikhel 2011), in which the vitellogenin (Vg) promoter was used to drive expression of a shortened version of GAL4, consisting of its DNA-binding and activation, but not the middle, domains. The Vg-GAL4 driver was inducible by a blood meal in females, which resulted in GFP expression in the fat bodies of females after blood feeding. In An. gambiae, the carboxypeptidase promoter was used to drive GAL4 expression in the midgut, visualized via UAS-eYFP reporter (Lynd and Lycett 2012). However, to achieve sufficiently high expression levels, the activation domain of GAL4 had to be replaced by the VP16 activation domain, which removed the ability of the modified GAL4 to bind GAL80, the repressor of the GAL4/UAS system. Another study on An. gambiae used the GAL4/UAS system to drive Cyp4g16/17-RNAi in oenocytes to affect production of cuticular hydrocarbons (Lynd et al. 2019). Random insertions of GAL4 were also used for enhancer-trapping in An. stephensi, resulting in six GAL4 driver lines with larval and/or adult expression in the fat body, midgut, maxillary palps, antennae, muscles, salivary glands, and other tissues (O'Brochta et al. 2012).
The Q-system is another binary expression system that functions similarly to GAL4/UAS but employs the qa gene cluster from the bread fungus Neurospora crassa (Potter et al. 2010; Riabinina et al. 2015; Riabinina and Potter 2016). It was also originally developed in Drosophila but turned out to work well across species from plants to vertebrates. In mosquitoes, it was first used to target Orco+ neurons of An. gambiae (Riabinina et al. 2016). The 9-kb putative promoter/enhancer region immediately upstream of the ATG translation start codon of Orco was polymerase chain reaction (PCR)-amplified and cloned into a piggyBac vector, followed by the QF2 transcription factor. The resulting expression pattern of QF2 faithfully labeled Orco+ neurons, and the Orco-QF2 driver has since been used to study the anatomy and function of the olfactory system in An. gambiae (Riabinina et al. 2016; Afify et al. 2019; Maguire et al. 2022). However, it proved very challenging to create other driver lines in mosquitoes by PCR-amplifying a putative enhancer/promoter region in the same manner (although see Ye et al. 2020). The intergenic sequences in mosquitoes are very large, and without an accurate method that predicts the location of an enhancer, it is impossible to pinpoint and PCR-amplify the enhancer. Instead, mosquito researchers turned to an alternative strategy that bypasses enhancer search. Matthews et al. (2019) used CRISPR–Cas9 to create a double-stranded break at the stop codon of the ppk301 gene, and by providing a DNA template for HDR, replaced the stop codon with a T2A–QF2 construct, creating a ppk301–QF2 driver line in Ae. aegypti. The QF2 was thus expressed in exactly the same pattern as the ppk301 gene under the control of endogenous ppk301 enhancer, the exact location of which is not known. QF2 and ppk301 were produced as two separate proteins because of the ribosome skipping the T2A peptide. This method has been repeatedly and successfully used in Ae. aegypti to study their sense of taste (Jové et al. 2020), olfaction (Shankar et al. 2020; Sorrells et al. 2021; Younger et al. 2020; Zhao et al. 2022), and the anatomy of their nervous system (Zhao et al. 2021). It has also been implemented in An. gambiae to make a chemosensory IR76b–QF2 driver line (Ye et al. 2021).
Both GAL4/UAS and the Q-system can efficiently drive transgene expression in various mosquito tissues; however, modifications of GAL4 may be required for stronger expression. The QF2 transactivator of the Q-system has been reliably strong, but some toxicity effects may be present for broad expression patterns (Basrur et al. 2020; Zhao et al. 2021). The weaker version of the QF2, named QF2w, may thus be the best choice for future Q-driver lines (Riabinina et al. 2015). In addition, QF2 or QF2w may be silenced by QS and reactivated by quinic acid—manipulations that work well in Drosophila (Riabinina et al. 2015) and C. elegans (Wei et al. 2012) but that have not yet been implemented in mosquitoes.
CRISPR–Cas9 GENE DRIVES
Overview of Gene Drives in Mosquitoes
Gene drives increase gene inheritance rates above Mendelian rates (super-Mendelian inheritance), allowing a genetic modification, the “payload,” to rapidly spread throughout mosquito populations. The use of gene drives in mosquitoes has attracted interest because of their potential to eradicate serious mosquito-borne diseases, such as malaria.
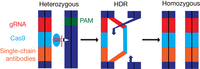
Gene drives were originally proposed in 2003 using naturally occurring homing endonuclease genes (HEGs) (Burt 2003). Although some HEG-based drives were produced, for example, in An. gambiae, I-SceI was used to disrupt GFP at an allele conversion (“homing”) rate of 56% (Windbichler et al. 2011), their development was limited because of the complexity of endonuclease engineering. The development of CRISPR–Cas9 as a tool for genome editing and the ease of engineering gRNAs originally facilitated the development of CRISPR–Cas9 gene drives (CGDs) in Drosophila (Gantz and Bier 2015). CGDs convert organisms heterozygous for a gene into homozygotes in an autocatalytic process termed mutagenic chain reaction (MCR) (Fig. 1; Gantz and Bier 2015; Gantz et al. 2015). Interestingly, a recent analysis by Garrood et al. (2021) in An. gambiae suggested that although CGDs can produce off-target mutations, these can be reduced to below detectable levels by tightly restricting spatiotemporal Cas9 expression to the germline and carefully choosing the guide RNA (gRNA) target site. Furthermore, there was no evidence that off-target mutations were capable of driving, even when using the drive vas-7280CRISPRh, which shows high rates of off-target mutations (Hammond et al. 2017; Garrood et al. 2021).
Mutagenic chain reaction (MCR) mechanism driving the first mosquito CRISPR–Cas9 gene drive (CGD) (Gantz et al. 2015). (Left) The drive construct contains sequences for gRNA (red), Cas9 (cyan), and Plasmodium falciparum–targeting single-chain antibodies (orange). Cas9 scans for the PAM sequence, and then gRNA checks for a complementary sequence. When the target sequence is found, the Cas9–gRNA complex cleaves (black and white arrow) the wild-type allele (dark blue). (Middle) The double-stranded break is repaired by homology-directed repair (HDR). (Right) Following HDR, the mosquito is homozygous for the drive construct.
Currently, mosquito CGDs are used for two general purposes: population suppression (reducing the number of mosquitoes) and population replacement (replacing a wild-type population with a modified one). For example, the population suppression drive vas-7280CRISPRh disrupted the An. gambiae female fertility gene AGAP007280. At peak drive frequency, this suppressed the egg production of a caged population by 92%. However, functional resistance alleles caused drive frequency to decline, making it unsuitable for field trials (Hammond et al. 2016, 2017). A population replacement drive was used to express single-chain antibodies targeting the malarial parasite P. falciparum (m2A10 and m1C3) in An. stephensi, showing inheritance rates of ∼99.5% (Fig. 1; Gantz et al. 2015). Although the efficacy was not tested, the coexpression of m2A10 and m1C3 has previously been shown to abolish P. falciparum development (Isaacs et al. 2012).
Resistance to Gene Drives
A significant challenge for CGDs is resistant alleles. In addition to natural genetic variation, CGDs can produce resistance alleles when double-stranded breaks are repaired by NHEJ pathways, producing indels that resist further cleavage (Esvelt et al. 2014). The protospacer-adjacent motif (PAM) sequence (Fig. 1) is particularly vulnerable to mutations, as a single-nucleotide mutation can prevent Cas9 function (Jinek et al. 2012). The PAM sequence is a short motif near the target sequence and is essential for Cas9 recognition of target DNA. Mathematical modeling suggests that resistant alleles will inevitably arise and spread through the population (Esvelt et al. 2014; Unckless et al. 2017), calling for approaches to reduce resistance development.
Approaches that reduce resistance rates may be combined to increase effectiveness. First, NHEJ rates can be reduced by tightly restricting drive expression to the germline, which shows higher rates of HDR (Gantz and Bier 2015). Previously, the vasa promoter has been used to drive Cas9 expression in An. stephensi (Gantz et al. 2015). However, leaky expression of Cas9 in female mosquitoes resulted in its maternal deposition into embryos, leading to resistance-generating NHEJ. Hammond et al. (2021a) found the zero population growth (zpg) promoter reduces leaky Cas9 expression and generates fewer resistance alleles in An. gambiae, significantly increasing drive inheritance, although functional resistance alleles were still able to emerge and reduce the drive prevalence (Hammond et al. 2021a). Inhibiting the NHEJ protein ligase IV by coexpressing the adenovirus4 proteins E1B55K and E4orf6 increases HDR efficiency by up to eightfold in mammalian cells (Chu et al. 2015), although this has not been tested in mosquitoes. However, suppressing NHEJ may not entirely prevent resistance-generation mutations, as HDR can produce errors (for review, see Guirouilh-Barbat et al. 2014).
Alternatively, targeting highly conserved sequences of essential genes can prevent the evolution of resistance. This has been showed in An. gambiae by targeting the female isoform dsxF of the sex determination gene doublesex (dsx). Any dsx mutations that may resist CGD action produce a nonfunctional dsxF transcript and are unable to spread (Kyrou et al. 2018; Simoni et al. 2020). The CGD produced by Kyrou et al. (2018) also recently became the first drive to be tested in larger cages containing age-structured populations with conditions closer to field conditions. Even in this setup, the drive reached 100% prevalence to crash the population (Hammond et al. 2021b). However, a recent drive targeting the An. gambiae female fertility gene AGAP029113 failed to spread in caged populations and was lost in three generations, despite heavy target sequence conservation. This was in part due to the emergence of a functional single-nucleotide substitution (Fuchs et al. 2021), demonstrating that targeting a conserved sequence is not always sufficient to prevent drive resistance.
For population replacement, essential genes could be targeted by a drive containing a rescue sequence that encodes the cognate protein. Mutated resistance alleles lacking this rescue sequence will experience negative selection due to the associated fitness cost, so will be unlikely to spread through the population (Esvelt et al. 2014). This has been showed by disrupting the kynurenine hydroxylase gene in An. stephensi (Adolfi et al. 2020). Resistant alleles lacking the rescue sequence significantly reduced female fecundity, which led resistant allele prevalence to decrease between generations. The drive reached >95% prevalence in five to 11 generations and remained stable for the following generations (tested up to 20 generations) (Adolfi et al. 2020).
Targeting multiple sites by multiplexing gRNAs will reduce resistance rates as it is unlikely that resistance alleles can form against each individual gRNA (Hammond et al. 2017; Marshall et al. 2017; Unckless et al. 2017). A recent population suppression CGD developed in Drosophila using four multiplexed gRNAs produced lower resistance and higher inheritance rates but plateaued at ∼63% population frequency and failed to suppress the population because of the drive fitness cost (Yang et al. 2022).
Controlling Gene Drive Spread
CGDs could potentially modify entire mosquito species, causing global effects. As such, it may be desirable to limit CGD spread. One approach to this is the split drive system, which allows a payload gene to be transiently driven to high frequency in a population while preventing global spread. In this system, drive elements are arranged so that each element drives the next, with no element able to drive itself. The top element will contain the payload gene (DiCarlo et al. 2015; Noble et al. 2019). Although this remains to be experimentally verified, the successive loss of each element of the chain over time would cause the payload gene to be diluted and eventually disappear from the population (Noble et al. 2019). Although split drives of three or more elements have yet to be developed, the transient nature of this system could theoretically be useful for field studies. However, mathematical modeling has suggested that split drives are likely to spread to neighboring populations, even at low migration rates. This is particularly true for population replacement approaches, which likely carry a lower fitness cost (Dhole et al. 2018).
Another approach that may reduce the invasive potential of gene drives is to unlink Cas9 from the other drive components. A proof-of-concept drive system using this approach has been developed in Ae. aegypti to disrupt the sex-linked white gene with the fluorescent marker tdTomato (Li et al. 2020b). Mathematical modeling suggests that this system will probably be reversible over time and less likely to invade neighboring mosquito populations. However, as Cas9 will not drive, this would require inundating the population with homozygous males, in contrast to other drives that require a single release (Li et al. 2020b).
When developing CGDs, strong biosafety procedures should be followed to prevent accidental release (Gantz and Bier 2015). In the case of an accident, one option could be to release mosquitoes resistant to CGD action, such as those carrying resistant alleles (Burt 2003; Unckless et al. 2017). This approach has not been implemented yet, but the occurrence of resistant alleles in previous drive experiments has decreased drive frequency (Hammond et al. 2017, 2021a; Simoni et al. 2020; Fuchs et al. 2021). This approach relies on a lower fitness cost of resistant alleles, so it could be an effective approach for population suppression drives, although it may be ineffective when targeting a highly constrained sequence or against population replacement drives (Taxiarchi et al. 2021). An alternative approach is releasing mosquitoes expressing anti-CRISPR effectors (e.g., the phage protein AcrIIA4), which inhibits both the PAM-binding site and RuvC endonuclease site of Cas9 (Dong et al. 2017). In caged populations with 50% of the mosquitoes carrying the sex-distortion drive produced by Kyrou et al. (2018), having 20% of the male An. gambiae express AcrIIA4 reduced drive inheritance to Mendelian rates. This caused drive frequency in a caged population to plateau at 76.6%–98.1% by the third generation. Egg production was maintained at 27%–43% of wild type by generation 16, preventing population collapse. However, if anti-CRISPR alleles are maintained in the population, this would likely prevent future CGD use (Taxiarchi et al. 2021).
Integral Gene Drives
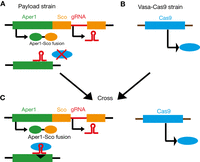
Integral gene drives (IGDs) could hypothetically be used to test population replacement approaches before spread by a drive. IGDs separately insert Cas9 and the payload gene into essential germline or somatic genes, respectively (Nash et al. 2019; Hoermann et al. 2021). The payload-carrying mosquitoes could be released for field trials. Cas9-carrying mosquitoes could be released later to cross with payload-carrying mosquitoes, allowing the payload to initiate driving (Fig. 2).
An example of a partial integral gene drive (IGD) developed by Hoermann and colleagues (2021). Separate mosquito strains were generated containing sequences for (A) the anti-Plasmodium effector peptide Scorpine (Sco, orange) fused to the somatic gene peritrophin1 (Aper1, green). Guide RNA (gRNA, red) is encoded by an artificial intron in Sco. As Cas9 (blue) is not present, the construct is inherited at Mendelian rates, allowing field testing in the absence of drive. (B) Cas9 (blue), driven by the vasa promoter (E. Marois, unpubl data). (C) Crossing the strains produces offspring carrying both the Sco payload and Cas9, allowing Sco to drive, as the homologous allele can be cleaved (black triangle) to initiate homing.
Hoermann and colleagues (2021) used a partial IGD strategy to drive an anti-Plasmodium peptide Scorpine (Sco) in An. gambiae. Sco was fused to the native somatic gene peritrophin1, expressed in the adult gut, and was driven by the native promoter. A separate vasa-Cas9 line was generated, in which Cas9 was driven by a vasa promoter but not associated with the native vasa gene (Fig. 2; Hoermann et al. 2021). IGDs can reduce the rate of evolution of functional resistant alleles, as frameshift-generation mutations could disrupt the native gene. However, nonframeshift mutations may still generate resistance alleles (Nash et al. 2019). This IGD gave homing rates of 98.43% and reduced infection intensity in mosquitoes fed P. falciparum. However, when Sco was linked to the amino-terminal of zinc carboxypeptidase A1, Plasmodium infectivity increased, demonstrating the importance of careful host gene choice (Hoermann et al. 2021).
FUTURE DIRECTIONS AND OUTLOOK
The development of genetic intervention in mosquitoes is strongly dictated by the practical needs to develop field applications and combat mosquito-borne diseases. RNAi applications and CGDs offer great promise here, although optimizing and rolling out these approaches to a wider range of species is required. For example, the efficiency of RNAi delivery via feeding may be increased by the down-regulation of larval gut RNases (Giesbrecht et al. 2020). To date, CGDs have mostly been produced for An. gambiae and An. stephensi (see Supplemental Table S3). CGD development has now begun in Ae. aegypti, although the drives developed so far have a lower drive efficiency (Li et al. 2020b). No CGDs have been produced for Culex mosquitoes, despite their epidemiological importance. However, early steps toward CGD development have begun in C. quinquefasciatus—expression toolkits for gRNA and Cas9 have been designed (Feng et al. 2021b) and gRNA target sites identified (Feng et al. 2021a). Interestingly, in C. quinequefasciatus, a gRNA scaffold variant containing a 5-bp loop that better represents the CRISPR–Cas native state increased allele conversion rates and improved drive efficiency when applied to Drosophila (Feng et al. 2021b), suggesting this could be applied to other species. Further research in other species could validate the efficacy of this loop variant and provide valuable information about the likely effectiveness of CGDs in eliminating a wide range of mosquito-borne diseases.
In addition to practical applications, recent interest in the basic biology of mosquitoes, such as their sensory neuroscience and sensory ecology, has driven the development of complementary genetic tools. Transgenic knock-ins and binary expression systems (see Supplemental Table S2) allow us to study neuronal morphology and function and to manipulate mosquito behavior. These approaches will also be applicable to mosquito species that do not transmit diseases but that are interesting because of their unusual biology and ecology.
ACKNOWLEDGMENTS
O.R. was funded by the Wellcome Trust (217440/Z/19/Z), the Royal Society (RGS/R2/192005), and the Natural Environment Research Council (NE/W00402X/1). M.Q. was funded by the BBSRC NLD DTP scholarship.
Footnotes
-
From the Mosquitoes collection, edited by Laura B. Duvall and Benjamin J. Matthews.
-
Supplemental material is available at www.cshprotocols.cshlp.org.