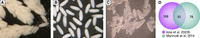Identification of Mosquito Eggshell Proteins from Aedes aegypti by Liquid Chromatography with Tandem Mass Spectrometry (LC–MS/MS) Proteomic Analysis
- 1Department of Entomology, The University of Arizona, Tucson, Arizona 85721, USA
- 2Analytical and Biological Mass Spectrometry Core Facility, The University of Arizona, Tucson, Arizona 85721, USA
- 3Department of Chemistry and Biochemistry, The University of Arizona, Tucson, Arizona 85721, USA
- ↵4Correspondence: jisoe{at}ag.arizona.edu
Abstract
The insect eggshell is a multifunctional structure with several important roles, including generating an entry point for sperm via the micropyle before oviposition, serving as an oviposition substrate attachment surface, and functioning as a protective layer during embryo development. Eggshell proteins play major roles in eggshell tanning and hardening following oviposition and provide molecular cues that define dorsal–ventral axis formation. Precise eggshell formation during ovarian follicle maturation is critical for normal embryo development and the synthesis of a defective eggshell often gives rise to inviable embryos. Therefore, simple and accurate methods for identifying eggshell proteins will facilitate our understanding of the molecular pathways regulating eggshell formation and the mechanisms underlying normal embryo development. This protocol describes how to isolate and enrich eggshells from mature oocytes of Aedes aegypti mosquitoes and how to extract their eggshell proteins for liquid chromatography with tandem mass spectrometry (LC–MS/MS) proteomic analysis. Although this methodology was developed for studying mosquito eggshells, it may be applicable to eggs from a variety of insects. Mosquitoes are ideal model organisms for this study as their ovarian follicle development and eggshell formation are meticulously regulated by blood feeding and their follicles develop synchronously throughout oogenesis in a time-dependent manner.
MATERIALS
Reagents
Acetonitrile
Acrylamide SDS–PAGE gels (12%; Bio Rad)
Adenosine triphosphate (100 mm stock)
Adult Aedes aegypti mosquitoes (e.g., Rockefeller, widely used strain) in a cage
-
Rear mosquitoes under standard laboratory conditions as previously described (Imam et al. 2014) (e.g., 28°C, 72% relative humidity, with a photoperiod of 16 h light and 8 h dark cycle)
Ammonium bicarbonate (50 mm, 100 mm)
Dithiothreitol (DTT; 25 mm) in 50 mm ammonium bicarbonate
Ethanol (90%, v/v, and 100%)
Formic acid
GelCode Blue Stain Reagent (Thermo Scientific)
Glacial acetic acid
Guanidine hydrochloride (6.0 m, pH 9.0)
H2O (boiling and room temperature)
Ice
Iodoacetamide (55 mm) in 50 mm ammonium bicarbonate
Methanol
Phosphate-buffered saline (PBS), pH 7.4
Pierce 660 nm protein assay reagent containing Ionic Detergent Compatibility Reagent (Thermo Scientific)
Solvent mixture for peptide extraction
Sucrose (10%, w/v, solution)
Tris–Glycine–SDS protein electrophoresis buffer
Ultrapure H2O
Whole vertebrate blood (e.g., whole bovine blood with citrate anticoagulant; HemoStat laboratories)
Equipment
Acclaim PepMap 100 trap column (75-µm inner diameter [ID] × 2-cm)
Acclaim PepMap rapid separation liquid chromatography (RSLC) analytical column (75-µm ID × 25-cm)
Bath sonicator
Centrifuge
Cotton balls
Dionex Ultimate 3000 RSLCnano System
Dounce homogenizer (5-mL-volume) with pestles A and B
Forceps
Gel staining box or large Petri dish (see Step 19)
Glass blood feeder (e.g., Chemglass CG-1836-75)
-
A Hemotek system (Hemotek Ltd.) could be used instead.
Glass Pasteur pipettes
Glass plate
Glass scintillation vials (20-mL)
Glass slide
Incubator
Lab clamp
Light microscope
Mass spectrometer equipped with a nano-flow liquid chromatography system
Mesh strainer (40-µm)
Microcentrifuge tubes (1.7-mL)
Microscissors (optional, see Step 7)
Mini gel electrophoresis unit
Mosquito cage
Parafilm
Petri dish, glass (100-mm × 15-mm)
Proteome Discoverer software
Razor blades
Refrigerator (4°C)
Scaffold Q + S software (Proteome Software Inc.)
Shaker table
SpeedVac concentrator
Surgical scalpels
Vortexer
Vortex tube adapter
Water bath
METHOD
-
Users must follow all institutional procedures for working with mosquitoes and seek guidance from the relevant regulatory bodies for such matters.
-
This protocol requires access to a mass spectrometer and extensive expertise with proteomic techniques, so users new to these approaches are encouraged to consult with relevant experts or a core facility.
Mosquito Eggshell Protein Extract Preparation
-
1. Set up glass blood feeders wrapped in stretched Parafilm, connected to a water bath (40°C), and place the glass feeders on the screened top of the mosquito cage, secured with a lab clamp. Prewarm vertebrate blood (e.g., cow) to 37°C.
-
2. Load 1 mL of prewarmed vertebrate blood supplemented with 5 mm adenosine triphosphate into the feeders according to the manufacturer's instructions.
-
3. Allow 3- to 4-d-old adult female mosquitoes in a cage to feed on the blood source for 1 h.
-
4. Cold-anesthetize mosquitoes by placing the cage for 5 min in a refrigerator (4°C).
-
5. Place 100 cold-anesthetized mosquitoes onto a glass Petri dish on ice under a light microscope to facilitate sorting of engorged mosquitoes based on abdominal distension (Fig. 1). Use forceps to transfer fully engorged mosquitoes to a new cage. Follow standard rearing protocols (see Protocol: Aedes aegypti Culturing and Egg Collection [Clemons et al. 2010]; Protocol: Batch Rearing Aedes aegypti [Wohl and McMeniman 2023]; Imam et al. 2014). Maintain blood-fed mosquitoes with 10% sucrose (via soaked cotton balls) for several days.
-
Although in general eggshell formation for each oocyte is completed ∼48 h after blood feeding, we recommend harvesting fully mature ovarian follicles at 4 d post–blood meal, which can be accomplished by depriving the gravid females of an oviposition substrate. Place the 10% sucrose-soaked cotton on the cage for only 1 h per day during this period to avoid mosquitoes ovipositing on the cotton.
-
-
6. On the day of dissection, cold-anesthetize mosquitoes as described in Step 4.
-
7. Place a drop of ice-cold PBS onto a glass slide. Place an anesthetized mosquito onto the drop of PBS. Under a light microscope, gently dissect ovaries from the mosquitoes by grasping the last two abdominal segments with forceps and pulling until the ovaries emerge (see Fig. 2 in Protocol: Visualization of Apoptotic Ovarian Follicles during Aedes aegypti Mosquito Egg Maturation by Fluorescent Imaging Studies (Isoe et al. 2023a).
-
If this technique is not successful because of the size of the fully developed ovaries, the dorsal side of the abdomen can be cut along the longitudinal axis with microscissors and the ovaries gently lifted out.
-
-
8. Using a glass Pasteur pipette, transfer ovaries into a glass scintillation vial containing 5 mL of ice-cold PBS (Fig. 2).
-
9. Once approximately 100 pairs of ovaries are collected in the vial, pipette up and down 20 times. As soon as the mature elongated oocytes settle to the bottom of the vial, use a glass Pasteur pipette to remove all floating tissues and cells including inner, lateral, and common oviducts; trachea; muscles; germarium; secondary follicles; and follicular epithelial cells. Add 5 mL of ice-cold PBS and repeat Step 9 10 times to ensure a minimum contamination of nontarget cell types.
-
Oocytes quickly descend because of gravity, whereas other ovarian tissues float or descend slowly.
-
-
10. Transfer the purified oocytes in 1 mL of ice-cold PBS into a Dounce homogenizer using a glass Pasteur pipette. Homogenize the oocytes on ice using 40 strokes in a Dounce homogenizer with pestle B. Let the insoluble eggshells form a pellet by gravity at the bottom of the glass homogenization tube. Remove the cloudy portion that contains vitellogenin yolk proteins and other soluble proteins with a glass Pasteur pipette. Add 1 mL of ice-cold PBS to resuspend the eggshell without homogenization, remove the cloudy portion, and repeat this process until the cloudiness disappears.
-
11. Transfer the homogenates containing eggshells onto a nylon mesh (40-µm) using a glass Pasteur pipette and wash thoroughly with 5 mL of PBS to remove oocyte cytosolic components. Repeat this washing process two additional times by pouring 5 mL of PBS onto the mesh to ensure a minimum contamination of cytosolic and membrane proteins (Fig. 2).
-
12. Add 1 mL of ice-cold PBS to the enriched eggshells on the mesh and quickly remove the PBS and eggshells with a glass Pasteur pipette. Transfer the enriched eggshells in PBS into a Dounce homogenizer using a glass Pasteur pipette. Remove the PBS and homogenize the eggshells in 5 mL of 6 m guanidine hydrochloride, pH 9 in a Dounce homogenizer with pestle A for 30 strokes. Place the homogenates in an incubator for 16 h at 37°C.
-
13. Precipitate the proteins by adding 45 mL of 100% ethanol and incubate the samples for 16 h at −20°C.
-
14. Pellet the eggshell proteins by centrifugation at 16,000g for 15 min at 4°C. Remove the supernatant carefully and wash the pellet with 90% ice-cold ethanol. Repeat the 90% ethanol wash two additional times to remove residual guanidine hydrochloride. Pellet the eggshell proteins by centrifugation at 16,000g for 15 min at 4°C after each wash.
-
15. Air-dry the pellets for 10 min to completely eliminate the ethanol.
-
16. Resuspend the eggshell proteins in 50 µL eggshell lysis buffer and use 2 µL of the resuspended protein to assay the protein concentration using the Pierce 660-nm reagent containing the Ionic Detergent Compatibility Reagent according to the manufacturer's instructions.
-
17. Add an equal volume of protein sample buffer to up to 20 µg of protein sample from Step 16 for a maximum of 50 µL and then denature the purified eggshell proteins in boiling H2O for 5 min.
A feeding status of individual mosquitoes observed under a light microscope. (A) An image of a 3-d-old sugar-fed mosquito. (B) An image of fully engorged mosquito taken 20 min after feeding bovine blood.
Representative images of dissected ovaries (A), enriched primary follicles (B), and isolated eggshell without cytosolic contents (C). A comparison of number of Ae aegypti eggshell proteins identified (D) between the method described here and in Isoe et al. (2023b) and the previous liquid chromatography with tandem mass spectrometry (LC–MS/MS) studies Marinotti et al. (2014).
Protein Sample Preparation for Mass Spectrometry
-
18. Load the mosquito eggshell protein samples onto an sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gel in Tris-Glycine-SDS protein electrophoresis buffer. Run the protein gels via a mini gel electrophoresis unit using standard techniques for ∼5 min until the proteins have entered the resolving gel (∼1 cm). This step is used to remove any nonprotein impurities.
-
19. Transfer the gel to a suitably sized gel staining box or large Petri dish and fix the gels with a 25-mL solution of 10% (v/v) glacial acetic acid and 50% (v/v) methanol for 30 min on a shaker table at room temperature. Stain the gel with 20 mL GelCode Blue Stain Reagent with shaking for 16 h at room temperature. Destain the gel using 25 mL ultrapure H2O three times for ∼2 h at room temperature on a benchtop orbital shaker. This step enhances stain sensitivity.
-
20. Excise gel bands (∼1.0 cm2) on a clean glass plate using a clean surgical scalpel and transfer the gel bands into 1.7-mL microcentrifuge tubes. Destain bands in 1 mL of 50% (v/v) methanol in 50 mm ammonium bicarbonate at room temperature on a vortexer with a tube adapter at low speed for 1 h or until the stain is removed; change the solution several times if necessary. Remove the destain.
-
21. To reduce disulfide bonds in the protein sample, add 100 µL of 25 mm DTT in 50 mm ammonium bicarbonate to the 1.7-mL microcentrifuge tube containing the gel bands for 45 min at 60°C. Quickly vortex the tube. Remove the DTT and immediately add 100 µL of 55 mm iodoacetamide in 50 mm ammonium bicarbonate for 30 min at room temperature to alkylate-free cysteines.
-
22. Remove the solution and wash the bands in 1 mL of 50 mm ammonium bicarbonate on a vortexer with a tube adapter or shaker for 1 h at room temperature. Repeat this wash two times.
-
23. Cut the bands into small pieces with a razor blade and transfer the bands into a 1.7-mL microcentrifuge tubes. Dehydrate the bands by adding 500 µL of 100% acetonitrile and incubating for 10 min at room temperature. Remove as much acetonitrile as possible. Dry the samples in a SpeedVac concentrator to remove the remaining acetonitrile.
-
24. Add 30 µL of eggshell protease cocktail to the samples. Incubate for 16 h at 37°C.
-
25. Centrifuge the gel pieces for 2 min at 4000g at room temperature, and transfer the supernatant to a fresh 1.7-mL microcentrifuge tube. Add 50 µL (or just enough to cover the gel pieces) of solvent mixture for peptide extraction to the remaining gel pieces. Sonicate for 10 min at room temperature. Centrifuge for 2 min. Transfer the supernatant to combine with the original supernatant.
-
26. Repeat the extraction in Step 25, transfer the supernatant to a new 1.7-mL microcentrifuge tube, pool the supernatants, and completely dry the supernatants in a SpeedVac concentrator for 30 min at room temperature.
LC–MS/MS and Protein Identification
-
27. Identify eggshell proteins by detecting tryptic/lysC peptides using a mass spectrometer equipped with a nano-flow liquid chromatography system followed by matching them to databases containing theoretical protein digestions.
-
In our laboratory, we use Thermo Scientific instruments, columns, and software. We have a Q Exactive Plus mass spectrometer incorporated with an EASY-Spray nanoESI source.
-
i. Resuspend peptides in 10 µL of 0.1% formic acid (1.0 µg/µL).
-
ii. Inject peptides (500 ng in 0.1% formic acid) onto an Acclaim PepMap 100 trap C18 Reversed Phase HPLC column, and elute peptides onto an Acclaim PepMap RSLC analytical column (separation column), using an H2O (0.1% formic acid) and acetonitrile (0.1% formic acid) gradient over 120 min.
-
iii. For MS, set the flow rate at 300 nL/min using a Dionex Ultimate 3000 RSLCnano System. Analyze peptides using a data-dependent method.
-
iv. For MS/MS, use a normalized higher collision energy set at 27 and an isolation width of 1.5 m/z.
-
v. Search MS and MS/MS raw data against the Ae. aegypti NCBI protein database and a common human-derived contaminant protein database using Proteome Discoverer software using a SEQUEST based search engine.
-
MS/MS spectra matches consider fully tryptic peptides with at most two missed tryptic cleavage sites. Variable post-translational peptide modifications are also considered (e.g., methionine oxidation and cysteine carbamidomethylation). To identify proteins at 95% confidence, use XCorr score cutoffs as determined by a reversed database search.
-
-
vi. Further analyze the peptide and protein identification outputs with proteomics software (e.g., Scaffold Q + S), a program that uses Bayesian statistics to detect additional spectra.
-
vii. Accept protein identifications that have at least two peptides at a 0.1% peptide false discovery rate with 95% protein confidence.
-
viii. Subject the identified eggshell proteins to BLASTP protein searches against the nonredundant NCBI protein database to find possible functions.
-
DISCUSSION
Eggshell proteins from Ae. aegypti mosquitoes were first identified and described more than 28 years ago (Lin et al. 1993; Edwards et al. 1998). When mosquito eggs are laid, a protective extracellular eggshell immediately undergoes final maturation by melanization and protein cross-linking reactions catalyzed by several eggshell enzymes. The key enzymes involved in these reactions for Ae. aegypti have been well identified and biochemically studied (Li 1994; Ferdig et al. 1996; Han et al. 2000; Johnson et al. 2001; Fang et al. 2002; Kim et al. 2005; Li and Li 2005). In addition, proteomic studies on purified mosquito eggshells have been conducted to identify novel eggshell protein components. The first proteomic study on the mosquito eggshell was performed in Anopheles gambiae in 2010 (Amenya et al. 2010). In Ae. aegypti, eggshell proteomic analysis identified 130 unique proteins (Marinotti et al. 2014). Traditionally, in these studies, a meticulous hand sorting of endochorion and exochorion after homogenization was performed under a light microscope to enrich and purify the eggshell before the extraction of eggshell proteins. Mosquito eggshell proteomics using the protocol described here have identified an additional 168 proteins in Ae. aegypti, indicating that a synthesis of mosquito eggshell is far more complex process than previously thought (Fig. 2; Isoe et al. 2023b). Some of these extracellular eggshell proteins may function together to rapidly engage in cross-linking and melanization steps upon oviposition to protect and support the embryonic development and the first larval instar during the extended periods of time these embryos may encounter before hatching.
The present protocol was developed to ensure a fast, accurate, and efficient separation of eggshells from dissected Ae. aegypti mosquito ovaries to subsequently identify eggshell proteins by LC–MS/MS analysis. Important features of this approach involve (1) an enrichment of primary oocytes that are surrounded by a protective eggshell from other components of the ovaries, (2) the physical separation of partially cross-linked extracellular eggshell from the oocytes by Dounce homogenization, (3) the extraction of eggshell proteins with the chaotrope guanidine hydrochloride, and (4) the subsequent LC–MS/MS analysis of mosquito eggshell proteins. The first step allows a thorough physical separation of the fully matured primary oocytes from other ovarian tissues and cell types including the inner, lateral, and common oviducts; trachea; longitudinal and circular muscles; germarium; secondary follicles; and follicular epithelial cells, without compromising the eggshell structure. We strongly recommend an enrichment of the primary oocytes as described herein, otherwise proteins from contaminating tissues will be identified as noise during downstream proteomic analysis. Thus, Step 9 should be thoroughly repeated until no unwanted tissues are visible in the vial under a light microscope, exclusively recovering primary oocytes covered with eggshell that can be used immediately in the next step. We chose Dounce homogenization in PBS on ice with pestle B in Step 10 to physically disrupt the primary oocytes to separate the eggshells from oocytes. Oocyte intracellular contents including vitellogenin yolk proteins become readily soluble and cloudy in buffer after homogenization, whereas insoluble eggshells quickly sediment to form a pellet at the bottom of the glass homogenization tube. This process is repeated following a removal of the top phase that includes a soluble fraction and nonsedimented cellular debris with a glass Pasteur pipette until the cloudiness disappears. The eggshell samples are filtered on nylon mesh (40-µm) and washed several times to remove remaining particulate and protein contaminants. The eggshell samples are then further homogenized with pestle A and treated with guanidine hydrochloride overnight to extract eggshell proteins. In summary, this isolation and sequencing approach can be used to quickly identify eggshell proteins from a variety of mosquito species, allowing us to greatly expand our understanding of eggshell biology in these important vectors of human disease (Fig. 2).
ACKNOWLEDGMENTS
We thank Carter J. Simington, Max E. Oscherwitz, and Alyssa J. Peterson at the University of Arizona for technical assistance. We thank Dr. Ning Zhang and reviewers for their valuable and thoughtful comments. Proteomic studies like those described are performed at the Analytical and Biological Mass Spectrometry Core Facility at the University of Arizona BIO5 Institute. Our research is funded internally through a Core Facilities Pilot Program provided by the Research, Innovation & Impact at the University of Arizona (to J.I.).
Footnotes
-
From the Mosquitoes collection, edited by Laura B. Duvall and Benjamin J. Matthews.