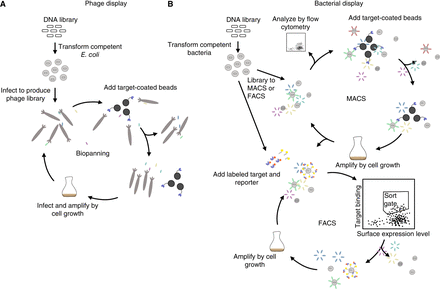
Common selection strategies for generating affibody molecules. Schematic overview of the selection processes in (A) phage display and (B) bacterial display. In phage display, the affibody library is displayed on bacteriophages and panned against the target coated on a solid support, such as paramagnetic beads. The binding population associated with the beads is isolated by washing away the nonbinding population. The binding population is amplified and the procedure is repeated two to four times to enrich for affibody variants that bind to the target. In bacterial cell display, the affibody library is displayed on the surface of bacterial cells and incubated with fluorescently labeled target and reporter molecules, followed by the isolation of binding populations using fluorescence-activated cell sorting (FACS). The binding population is amplified, and the procedure is repeated two to four times to enrich for affibody variants that bind to the target. Note that, for larger libraries, magnetic-activated cell sorting (MACS) is advisable to reduce the library sizes to allow efficient FACS.










