Study of Dendrite Differentiation Using Drosophila Dendritic Arborization Neurons
- Jason Y. Tann1,
- Fangke Xu1,
- Minami Kimura1,
- Oliver R. Wilkes1,2,
- Li-Foong Yoong1,
- Henrik Skibbe3 and
- Adrian W. Moore1,4
- 1Laboratory for Neurodiversity, RIKEN Center for Brain Science, Wako-shi, 351-0106, Japan
- 2Department of Cellular and Molecular Biology, Institute for Translational Medicine, University of Liverpool, Liverpool L69 3BX, United Kingdom
- 3Brain Image Analysis Unit, RIKEN Center for Brain Science, Wako-shi, 351-0106, Japan
- ↵4Correspondence: adrian.moore{at}riken.jp
Abstract
Neurons receive, process, and integrate inputs. These operations are organized by dendrite arbor morphology, and the dendritic arborization (da) neurons of the Drosophila peripheral sensory nervous system are an excellent experimental model for examining the differentiation processes that build and shape the dendrite arbor. Studies in da neurons are enabled by a wealth of fly genetic tools that allow targeted neuron manipulation and labeling of the neuron's cytoskeletal or organellar components. Moreover, as da neuron dendrite arbors cover the body wall, they are highly accessible for live imaging analysis of arbor patterning. Here, we outline the structure and function of different da neuron types and give examples of how they are used to elucidate central mechanisms of dendritic arbor formation.
BACKGROUND
The functional connectivity of a dendrite arbor is delineated by its morphology. Global parameters such as arbor field size and branch density control input number, whereas local dendrite targeting regulates the position and identity of these inputs. In addition, the arbor function is modified by regionalized branch topologies that facilitate computation within branches and at branch points (Branco and Häusser 2010; Lefebvre et al. 2015; Poirazi and Papoutsi 2020).
Arbor morphology is created through dendrite targeting, branching, and growth during differentiation. Both intrinsic programs and an extracellular environment of signaling and biomechanical cues regulate these pattern-formation processes (Lefebvre et al. 2015). Significant data on the control of dendrite arbor differentiation have been derived from studies of neurons in culture, as this experimental approach restricts environmental parameters and allows their manipulation (Schelski and Bradke 2017). However, the unique physical and chemical properties of the in vivo environment cannot be completely represented in culture, and these can also be critical regulators of the differentiation process. In addition, the extent and nature of both intrinsic and extrinsic developmental controls are constantly changing as neurons go through a coordinated series of differentiation steps to create the final dendritic arbor structure (Yoong et al. 2019). Because of these extrinsic and temporal factors, it is also important to study arbor differentiation in its unique in vivo context (Tavosanis 2021).
Developmental and cell biological analyses of dendrite formation aim to uncover the fundamental mechanisms of cell growth and patterning that create the complex arbor. These studies also allow us to examine how abnormalities in the arbor arise and lead to pathology in neurodevelopmental disorders (Kulkarni and Firestein 2012; Martínez-Cerdeño 2017; Forrest et al. 2018). In addition, they help to build computational models linking neuron pattern creation to mature function (Poirazi and Papoutsi 2020).
MORPHOLOGICAL AND FUNCTIONAL DIVERSITY OF DENDRITIC ARBORIZATION NEURON DENDRITE ARBORS
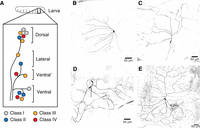
Dendritic arborization (da) neurons are a subset of peripheral sensory neurons. In the larva, investigators commonly examine the 15 da neurons of each abdominal segment, which are divided into four subtypes, named from class I to class IV (usually shortened to c1da–c4da) in order of increasing dendrite arbor morphological complexity (Table 1; Fig. 1; Grueber et al. 2002). The dendrite arbor morphologies of da neurons are easily assayed because these dendrites are located on the internal surface of the body wall epithelium and therefore accessible for live imaging (see Protocol: Mounting of Embryos, Larvae, and Pupae for Live Drosophila Dendritic Arborization Neuron Imaging [Xu et al. 2023a]). They can also be examined by filleting the larva with subsequent immunostaining (see Protocol: Filleting and Immunostaining of Larvae to Visualize Drosophila Dendritic Arborization Neuron Dendrite Arbors [Xu et al. 2023b]). Dendrite arbor shape parameters (also called morphometrics) are used to classify the neurons (Grueber et al. 2002) and to quantitate the action of dendrite arbor differentiation programs (Ferreira Castro et al. 2020). The number and position of the dendrite branch termini were key in initially classifying the da neuron subtypes and are critical to measure the function of arbor differentiation programs; the freely available software DeTerm is optimized to measure these parameters (Kanaoka et al. 2019) (see Protocol: Use of DeTerm for Automated Drosophila Dendrite Arbor Terminal Counts [Kimura et al. 2023]). On top of morphology parameters, neuron functional parameters and transcriptome identity are critical to determine a neuron subtype (Yuste et al 2020). The classification of da neuron subtypes by hallmark dendrite arbor morphologies correlates with their differences in axon targeting (Grueber et al. 2007), their sensory modalities, and their transcriptional controls.
C1da–c4da neurons in the Drosophila larva. (A) Drosophila dendritic arborization (da) sensory neurons in the abdominal hemisegment form a stereotypic arrangement. They can be identified by position, morphology, and markers. (B–D) Mosaic analysis with a repressible cell marker (MARCM) clones of c1da–c4da neurons; these clones were derived as described in Protocol: Mosaic Analysis with a Repressible Cell Marker (MARCM) Clone Generation in da Neurons [Tann et al. 2023b]. B is a ddaD c1da neuron, C is a ddaB c2da neuron, D is a ddaA c3da neuron (left soma) with an external sensory (ES) neuron (right soma) to the side, and E is a ddaC c4da neuron. (B–D were provided by Saori Akimoto.)
Drosophila dendritic arborization (da) neuron identities in each cluster
C1da neurons have the least complex branching morphology. Their branches are organized with a well-spaced comb-like topology. C2da neurons have longer dendrites but still have low overall arbor complexity. C3da neurons have higher arbor complexity. They have long primary and secondary branches, which are decorated with actin-rich interstitial protrusions, sometimes termed short terminal branchlets (STBs) (Andersen et al. 2005; Nagel et al. 2012). C4da neurons possess the most complex branching morphology, with repetitive bifurcations of the major branches. Their dendrite arbors extend until the terminal branches meet those of neighboring c4da neurons. This organization is known as tiling, which is the contiguous and nonoverlapping arrangement of arbors, often among different neurons of the same functional subtype (Grueber and Sagasti 2010; Lefebvre et al. 2015).
C1da neurons receive mechanosensory input that reports body wall contraction, acting as proprioceptors, which are sensory receptors that respond to body position or movement, to control larval locomotion. To do this, they use the mechanosensitive transmembrane channel-like (Tmc) protein. A combination of direct observation of calcium influx during larva locomotion and computational modeling shows how their comb-like arbor topology contributes to their proprioceptive function. Their secondary branches align along the body in an anterior–posterior orientation, which is the direction of body contraction during crawling. This branch orientation allows for maximal membrane curvature during contraction, which is hypothesized to directly lead to the opening of the mechanically gated Tmc channels (Cheng et al. 2010; Hughes and Thomas 2007; Guo et al. 2016; He et al. 2019; Vaadia et al. 2019; Ferreira Castro et al. 2020). At the ultrastructural level, c1da dendrites have a dense interlinked microtubule arrangement and tight tethering at specific sites of the body wall. Such an organization is likely linked to their proprioceptive functions, as similar interlinked microtubule cell ultrastructure is also seen in the other common type of Drosophila proprioceptive organs, known as the chordotonal organs (Dettman et al. 2001; Delandre et al. 2016).
C2da and c3da neurons participate in light touch responses by using the transient receptor potential cation channel (TRP) no mechanoreceptor potential C (NompC) and the polycystin cation channel (PCC) brivido-1 (Brv1) (Tsubouchi et al. 2012; Yan et al. 2013; Zhang et al. 2018). It has been suggested that the mechanosensory functionality of c3da neurons uses the STB structures specific to this subtype (Tsubouchi et al. 2012). C3da neurons are polymodal as, in addition to sensing light touch, these neurons also act as cold sensors. Cold sensing is mediated by NompC as well as transient receptor potential melastatin (Trpm), Brv1, and the TRP/PCC polycystic kidney disease 2 (Pkd2) (Turner et al. 2016, 2018).
The c4da neurons are also polymodal; they sense different stimuli through different receptors. Thermal and chemical stimuli are sensed through the TRP channels Painless (Pain) and TrpA1, whereas mechanical stimuli are sensed through the Piezo ion channel family factor Piezo and the Degenerin/epithelial sodium channel (DEG/ENaC) family channels Pickpocket1 (Ppk) and Pickpocket26 (Ppk26) (Tracey et al. 2003; Zhong et al. 2010; Kim et al. 2012b; Gorczyca et al. 2014; Mauthner et al. 2014). Finally, c4da neurons sense ultraviolet, violet, and blue light through the G-protein-coupled receptor Gr28b (Xiang et al. 2010; Yamanaka et al. 2013). There is evidence that the extensive and tiled dendrite arbor organization of c4da arbors may contribute to sensitivity and acuity. Reduced c4da arbor structures exhibit reduced sensitivity to thermal nociception, and hyperbranched arbors show hypersensitivity (Honjo et al. 2016).
During pupation, some larval da neurons are lost, leaving around six in the adult abdominal segments, with segment–segment variation (Shimono et al. 2009). In the neurons that remain, dendritic pruning and subsequent regrowth occur (we refer the reader to a recent in-depth review on this topic by Furusawa and Emoto 2021). This transforms the dendrite arbors of larval da neurons into adult ones. The regrowth of adult arbors provides an excellent system for in vivo imaging studies of arbor growth, in part because the larger size of the neurons (in comparison to those of the embryo and larva) allows for the precise and careful deconstruction of subcellular trafficking and cytoskeletal-reorganization events (Yoong et al. 2020). In the adult, c4da neurons again rebuild their extensive, highly branched topology. However, some other neuron subtypes rebuild arbors with substantial differences in morphology from their larval stages. For example, the c1da ddaE neurons undergo a dramatic increase in branching when regrowing for the adult stage (Williams and Truman 2004). Despite these discoveries, the functional roles of the adult da neurons are undescribed (Montell 2021).
TRANSCRIPTIONAL CONTROL OF SUBTYPE-SPECIFIC DENDRITE ARBOR MORPHOLOGY
More than a century ago, Santiago Ramón y Cajal beautifully illustrated the tremendous diversity in dendrite arbor morphologies (Ramón y Cajal 1911). This diversity arises because arbor-development programs are regulated as part of neuronal fate control, and different arbor morphologies of different neuron subtypes are specified through the action of transcription factors (Pai and Moore 2021). Genome-wide screens and in-depth single-candidate analyses have linked the action of various transcription factors to defining the hallmark morphological features of the different da neuron subtypes (Moore et al. 2002; Grueber et al. 2003a; Li et al. 2004; Sugimura et al. 2004; Kim et al. 2006; Parrish et al. 2006; Hattori et al. 2007; Jinushi-Nakao et al. 2007; Crozatier and Vincent 2008; Karim and Moore 2011b; Ferreira et al. 2014; Wang et al. 2015; Corty et al. 2016; Sears and Broihier 2016; Das et al. 2017).
One example of a transcription factor-driven neuron-subtype-specific morphogenesis program is the function of the BTB-ZF (Broad complex, Tramtrack, and Bric à brac-Zinc Finger) transcription factor Abrupt, which is expressed only in c1da neurons (Li et al. 2004; Sugimura et al. 2004). Abrupt modulates the levels of the γ-TuRC-tethering protein Centrosomin. Centrosomin orients microtubule polymerization at the tips of growing branches in the differentiating dendritic arbor to suppress branching frequency (Yalgin et al. 2015). This keeps the arbor structure of the c1da neuron simple. Abrupt also up-regulates the expression of Tenurin-m, which is a cell–cell adhesion molecule. This results in increased Tenurin-m expression along the c1da branches. da neurons elaborate dendrites in an approximately two-dimensional territory across the basal surface of the body wall epidermis. Tenurin-m is also expressed in the epidermis in a gradient, with high expression in the cells of the center and low expression in the cells at the boundaries of each hemisegment. Homophilic interactions between Tenurin-m in the epithelium and in the branches orient the growth of the branches along this gradient, which shapes the comb-like arbors that support c1da function (Hattori et al. 2013).
Another example is the role of the CUT homeodomain factor Cut, which lies at the center of a transcription factor network that defines arbor morphology differences between da neuron subtypes. C2, c3, and c4da neurons express Cut at different levels, with c2da neurons expressing the least and c3da neurons expressing the most. Higher Cut levels increase arbor complexity, and in c3da neurons, Cut also promotes the formation of STBs (Grueber et al. 2003a; Nagel et al. 2012). Cut exerts its effect by suppressing the expression of the POU (Pit-1, Oct-1, Unc-86) transcription factors Pdm1 and Pdm2. These factors are expressed in non-da peripheral nervous system neurons, where they suppress arbor branching. Cut prevents Pdm1 and 2 from being expressed in dorsal and lateral cluster c2da and c3da neurons, therefore enabling branching. In c2da neurons, Cut protein levels must be kept low, and, in the dorsal cluster c2da neuron ddaB, this is accomplished through the action of the TEA (TEF-1 and abaA) transcription factor Scalloped. If Scalloped (or its partner Vestigial) is lost from ddaB, Cut levels increase, and the cell is transformed into a c3da-like neuron, complete with STBs (Corty et al. 2016). Cut expression is higher in c4da neurons than in c2da neurons, and this increased cut expression is promoted by the BTB-ZF transcription factor Longitudinals lacking (Ferreira et al. 2014).
The arbor of c4da neurons is both larger and more complex than that of c3da neurons. Achieving this requires the combinatorial action of Cut and the early B cell factor (EBF) Knot (Hattori et al. 2007; Jinushi-Nakao et al. 2007; Crozatier and Vincent 2008). Knot promotes additional outgrowth and branching on top of that driven by Cut, in part because it promotes expression of the microtubule severing AAA ATPase Spastin. Knot also suppresses the ability of Cut to create STBs, which helps shape the differences in terminal branch morphology between c3da and c4da neurons (Jinushi-Nakao et al. 2007). Knot regulation of c4da dendrite arbor morphology also works through regulating Tenurin-m, which in turn helps to orient the terminal branches (Hattori et al. 2013).
DENDRITE BRANCH FORMATION
Studies in culture have begun to build a picture of the cell biology of neuronal arbor differentiation. Although Drosophila is not a traditional organism in which to examine these processes in culture, it is useful because of its genetic tractability (Sánchez-Soriano et al. 2010), and da neurons can be cultured to examine the stages of dendrite arbor specification and branching (Yoong et al. 2020; see Protocol: Culture of Larval and Pupal Drosophila Dendritic Arborization Neurons [Tann et al. 2023a]). Nevertheless, as the in vivo environment differs significantly from that in culture, the field also needs experimental models in which investigators can examine differentiation processes in situ (Tavosanis 2021). da neurons are a powerful model with which to do this. This is in part because of the large range of genetic tools available in Drosophila to manipulate neurons, including easy tools to produce mutant da neuron clones that allow the study of embryonically lethal genes and the determination of cell autonomy for a mutant phenotype, see Protocol: Mosaic Analysis with a Repressible Cell Marker (MARCM) Clone Generation in Drosophila Dendritic Arborization Neurons (Tann et al. 2023b) and also Karim and Moore (2011a). It is also because the effects of mutations on the arbor are easily assayed by immunohistology or live imaging, see Protocol: Filleting and Immunostaining of Larvae to Visualize Drosophila Dendritic Arborization Neuron Dendrite Arbors (Xu et al. 2023b) and Protocol: Mounting of Embryos, Larvae, and Pupae for Live Drosophila Dendritic Arborization Neuron Imaging (Xu et al. 2023a). Because of their experimental strengths, da neurons have been used to examine key cell biological mechanisms involved in dendrite differentiation—for example, microtubule motor procession, autophagy and secretory pathways, and exocytosis and endocytosis processes (Ye et al. 2007; Satoh et al. 2008; Zheng et al. 2008; Yang et al. 2011; Iyer et al. 2013; Arthur et al. 2015; Peng et al. 2015; Clark et al. 2018). In a series of studies, they have also been used to dissect mechanisms of dendrite branch formation.
In vivo time-lapse procedures have been used to dissect the local dynamic cytoskeletal-reorganization cascades that underlie da neuron dendrite branch initiation and growth (Andersen et al. 2005; Dimitrova et al. 2008; Nagel et al. 2012; Ori-McKenney et al. 2012; Stewart et al. 2012; Yalgin et al. 2015; Nithianandam and Chien 2018; Stürner et al. 2019; Tang et al. 2020; Yoong et al. 2020), see Protocol: Mounting of Embryos, Larvae, and Pupae for Live Drosophila Dendritic Arborization Neuron Imaging (Xu et al. 2023a). These live imaging approaches have used fluorescent protein-tagged markers to examine actin organization. They have shown how two types of actin puncta appear before interstitial dendrite branch initiation (Andersen et al. 2005; Nithianandam and Chien 2018; Stürner et al. 2019). The first, named actin blobs, are a type of puncta that migrate to the position of branch formation and then stall and contribute to branch initiation (Nithianandam and Chien 2018). The second type of puncta, named actin patches, form at branch formation sites through local F-actin nucleation and transform into filopodia (Andersen et al. 2005; Stürner et al. 2019). Genetic manipulation of actin regulators has provided a model of branch initiation at these sites. Activation of the Rho family GTPase, Rac1, is required to initiate actin patch formation. Downstream from this, the major actin regulator complex, Arp2/3, mediates nucleation of bifurcated actin polymers, which occurs through another actin cytoskeleton regulatory complex that controls cytoskeletal dynamics, known as WAVE/WASP complex. Arp2/3 and WAVE are commonly implicated in the ruffling of actin-based membrane protrusions, and here, they initiate the outgrowth of the nascent branch (Stürner et al. 2019). In c3da neurons, interstitial branches transform into STBs through the invasion of Fascin and CamKII, which regulate the rate of actin turnover within the structures, while dendrite branch stabilization in c4da neurons requires the invasion of the nascent branches by microtubules (Andersen et al. 2005; Moore 2008; Nagel et al. 2012).
A range of studies in da neurons has shown how terminal branch stabilization requires microtubule nucleating, polymerizing, and severing factors (Jinushi-Nakao et al. 2007; Ori-McKenney et al. 2012; Stewart et al. 2012; Nguyen et al. 2014; Yalgin et al. 2015; Tang et al. 2020). da neurons have been used extensively to investigate the issue of where microtubules are nucleated in dendrites (we refer the reader to a recent in-depth review on this topic by Wilkes and Moore (2020)). Studies conflict about the role of dendritic Golgi fragments (Golgi outposts) and also show dendritic endosomes to be microtubule-nucleation sites (Ori-McKenney et al. 2012; Zhou et al. 2014; Yalgin et al. 2015; Mukherjee et al. 2020; Nye et al. 2020; Weiner et al. 2020; Yang and Wildonger 2020).
High-resolution time-lapse imaging of growing pupal c4da neurons was used to study the process of main arbor branch formation. This study took advantage of the higher subcellular spatial resolution in the differentiating pupal dendrites compared to that of the embryo or larvae. It revealed a mode of actin regulation that guides microtubule polymerization into nascent branches. Tip branching is preceded by the up-regulation of anterograde microtubule activity at the dendrite tip in a widened growth cone (called a dendrite-partitioning module). Within this structure, the actin tails of filopodia extend toward a site of tip microtubule generation. These tails capture polymerizing microtubules, and repetitive capture events by the same tail guide concerted microtubule growth into a specific filopodia. This, in turn, transforms that filopodia into a branch. The amount of branch formation is controlled by the degree of actin tail extension, which is controlled by the atypical actin motor Myosin 6 (Yoong et al. 2020).
A complementary approach to local time-lapse imaging is to describe the outcome of genetic manipulations by creating statistical representations of global arbor cytoskeletal organization. This is done by measuring fluorescent actin- and microtubule-associated markers to report the local density of cytoskeletal features and then replicating these detailed measurements arbor-wide (Ferreira et al. 2014; Nanda et al. 2018, 2020).
DENDRITE SELF-AVOIDANCE AND TILING
Interactions between dendrites generate evenly covered territories (Grueber and Sagasti 2010; Lefebvre et al. 2015). Iso-neuronal (self) interactions lead to avoidance events that allow sister branches of the same neuron to spread out without much overlap within their own arbor. Hetero-neuronal interactions regulate how the arbor of one neuron is shaped by the position of the arbor of its neighbors, and this commonly occurs between neurons of the same subtype.
Dendritic iso-neuronal avoidance is a feature of all da neurons. To achieve this, dendrite branches from the same neuron subtype must recognize when they are touching. This suggests the action of a cell surface molecule. Disruption of the seven-pass transmembrane cadherin Flamingo and the immunoglobulin superfamily (IgSF) protein Turtle results in a mild breakdown of da dendritic self-avoidance (Long et al. 2009; Matsubara et al. 2011). However, given the large number of neurons existing in the same space, a recognition process for each individual neuron to detect self versus non-self entities may require extensive recognition diversity (Kramer and Stent 1985). For da neurons, this diversity is provided by Down syndrome cell adhesion molecule 1 (Dscam1), an IgSF transmembrane adhesion molecule that exists in more than 38,000 isoforms because of alternative splicing in its extracellular and transmembrane regions (Schmucker et al. 2000). This splicing diversity means that each da neuron expresses its own unique set of Dscam1 isoforms, which allows it to distinguish its own branches upon a branch–branch meeting during growth. When Dscam1 is removed, dendrites of the same neuron no longer repel each other during growth, leading to extensive branch fasciculation or overlap. In contrast, if two different da neurons that have dendrites that normally coexist in the same space are manipulated to both express the same Dscam1 isoform, their dendrites now repel (Hughes et al. 2007; Matthews et al. 2007; Soba et al. 2007). The iso-neuronal avoidance mediated by Dscam1 further acts in the context of its local growth environment to shape arbor morphology. This is seen in c3da neurons, in which Dscam1-mediated self-repulsion acts to temper the effects of dendritic attraction signals provided by locally secreted Netrin (Matthews and Grueber 2011).
Hetero-neuronal avoidance is shown by two da neuron subtypes. C4da neurons undergo tiling, and in studies in which c3da neurons are present at supernumerary levels, this neuron type also tiles (Grueber et al. 2003b). As with other aspects of arbor-pattern formation, the ability to tile is ultimately defined by the transcriptional cascades that specify neuron subtype (Karim and Moore 2011b). Because tiling requires interaction between dendrites of different neurons at the local level, it is predicted to involve molecules related to cellular contacts between dendrites. Surprisingly, while there is some evidence that Flamingo plays a role (Gao et al. 2000; Kimura et al. 2006), a clear candidate molecule underlying tiling has yet to be discovered (Grueber and Sagasti 2010).
In addition to interactions between dendrites, the physical placement of da dendrites is necessary to support their tiling. da neurons elaborate dendrites in a mostly two-dimensional territory across the basal surface of the epidermis and below an overlaying basal membrane (BM). This is a complex environment where multiple signaling and mechanical processes work in concert to regulate local branch outgrowth and density (we refer the reader to a recent in-depth review on this topic by Yin et al. 2021). In this environment, integrins expressed by the neurons mediate the attachment of dendrites to the BM (Han et al. 2012; Kim et al. 2012a; Jiang et al. 2014). Sema-2b/PlexB signaling activates Tricornered, an NDR kinase family member, within the neurons to also promote dendrite-to-BM adhesion (Meltzer et al. 2016). To do this, Tricornered acts in a pathway with Furry and with the target of rapamycin (TOR) complex factors TORC2, Rictor, and Sin1 (Emoto et al. 2004; Koike-Kumagai et al. 2009). Disruption of processes regulating dendrite-to-BM adhesion change not only hetero-neuronal but also iso-neural repulsion.
When the c4da branches are not attached to the BM, they are engulfed by the underlying epithelial cells via a program initiated by the localization of the phospholipid PIP2 to the epithelial surface adjacent to the dendrite. This is then followed by the recruitment of Rho1 to drive epithelial cell cortical F-actin remodeling (Jiang et al. 2019). The engulfed dendrites then become enclosed in adherens junction (Armadillo, E-cadherin) and septate junction (Coracle, Discs large, Neurexin, Neuroglian, and Scribble) proteins (Kim et al. 2012a; Tenenbaum et al. 2017; Yang et al. 2019). In addition to changing interactions for dendrite repulsion, these dendrite-engulfment structures restrict local dendrite growth and branching.
DIFFERENT MODES OF DENDRITE ARBOR GROWTH
Neurons undergo two phases of growth. First, there is a rapid early growth phase when a nascent neuron establishes its initial dendritic coverage. C1da neurons reach their mature arbor pattern by late embryonic stages, but c4da neurons continue to undergo pattern formation into early larval stages (Sugimura et al. 2003; Parrish et al. 2009; Yalgin et al. 2015; Ferreira Castro et al. 2020; Palavalli et al. 2020). Second, as an animal grows, the established neurons must expand their dendrite arbor to sustain proper connectivity (Yoong et al. 2019). After hatching, Drosophila larvae rapidly grow, undergoing an ∼200-fold gain in mass within a few days (Church and Robertson 1966), and subsequently, a corresponding extensive expansion of c4da arbors is required to maintain their dense tiling coverage. This extensive growth relies on fatty acid production, which is controlled at the transcriptional level by the sterol regulatory element-binding protein SREBP (Meltzer et al. 2017; Ziegler et al. 2017). It also requires amino acid transport into the neurons via the transporter Pathetic (Lin et al. 2015).
In the second growth phase, an isometric scaling mechanism coordinates arbor growth in proportion to larva body wall expansion. For c4da arbors, this depends on local epidermal-derived signals. The microRNA bantam acts in the epithelial cells reducing Akt-mediated growth activity in the adjacent neuron. Bantam also progressively increases levels of integrin on the surface of the epithelial cells, and this binds to the BM, which displaces the dendrites from the BM and increases the interaction between the epithelial layer and the dendrites to couple the rate of dendrite expansion to that of the body wall (Parrish et al. 2009; Jiang et al. 2014).
DEVELOPMENTAL RULES FOR DENDRITE ARBOR PATTERN FORMATION
Because dendrite arbor differentiation goes through a series of distinct steps, to understand how the final arbor pattern is achieved, investigators aim to follow arbor formation through all these steps (Yoong et al. 2019). Recent development of long-term in vivo imaging to follow da neurons during arbor differentiation now provides a way to do this (Ferreira Castro et al. 2020; Palavalli et al. 2020; Yoong et al. 2020; see Protocol: Mounting of Embryos, Larvae, and Pupae for Live Drosophila Dendritic Arborization Neuron Imaging [Xu et al. 2023a]).
The comb-like structure of the c1da neuron vpda dendrite arbor is used in the mechanical sensing of body wall contraction in proprioception (He et al. 2019; Vaadia et al. 2019; Ferreira Castro et al. 2020). Two studies used quantitative in vivo imaging to track the formation the arbor vpda; the authors then drived a set of branching rules that govern arbor pattern formation. The main primary branch grows out, followed by a phase of secondary branch addition. This is followed by subsequent branch addition of higher-order branches by repeated cycles of extension and retraction. A branch retraction and removal process then follows, which is hypothesized to minimize the total amount of material used in the arbor. Following this is a period of stabilization. This cycle drives the basic process of neuron patterning, and one study showed that sequencing first growth and then stochastic retraction is sufficient to generate vpda arbor topology (Ferreira Castro et al. 2020). A second study described that the branch elimination process is triggered upon dendrite–dendrite iso-neural contact through the action of Dscam1, and the retraction of branches occurs only until the prior branchpoint. This mechanism biases the elimination of branches that have a higher probability for dendrite-dendrite contact, which selectively removes terminal (or final-order) branches, limits local branch density, and enriches for the retention of branches that form perpendicular to their parent (Palavalli et al. 2020). Ultimately, the application of these rules results in the comb-like structure of vpda.
SUMMARY
The connectivity of neurons is regulated through transcriptional programs that define global arbor morphology parameters and local cell remodeling mechanisms of arbor differentiation. Here we have highlighted how da neurons have been used to examine the genetic logic of transcriptional control. We have discussed how work in these neurons has elucidated essential cell–remodeling molecular mechanisms of dendrite growth, branching, self-avoidance, and tiling. In addition, we have reviewed how live imaging and mathematical modeling with these neurons have revealed the developmental rules that combine these remodeling processes to create specific arbor morphologies.
ACKNOWLEDGMENTS
J.Y.T. is supported by a Japan Society for the Promotion of Science Foreign Postdoctoral fellowship (Japan). O.R.W. is supported by an International Program Associate fellowship jointly administered by RIKEN (Japan) and the University of Liverpool (United Kingdom). F.X. is supported as a RIKEN Special Postdoctoral Researcher (Japan). L.-F.Y. was supported by a Human Frontier Science Program Long-Term Fellowship. We thank Yang Xiang for feedback on the manuscript and Saori Akimoto for images used in this article.
Footnotes
-
From the Drosophila Neurobiology collection, edited by Bing Zhang, Ellie Heckscher, Alex C. Keene, and Scott Waddell.