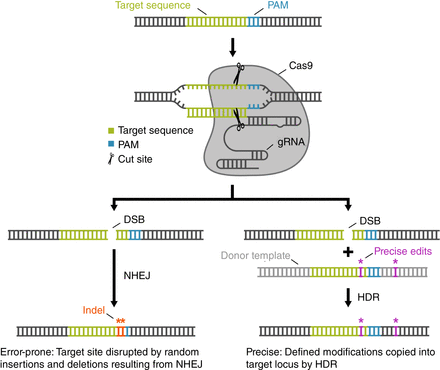
Gene editing with CRISPR–Cas9: components and DNA-repair pathways. The CRISPR–Cas9 gene-editing system uses two simple components to induce targeted double-stranded breaks (DSBs), the repair of which provides an opportunity to edit the genome. (Top) The Cas9 enzyme cleaves both strands of DNA using its two nuclease domains and a guide RNA (gRNA). As its name implies, the gRNA “guides” Cas9 to its target site, which must be adjacent to a 3′ protospacer-adjacent motif (PAM) for cleavage to occur. The widely used Streptococcus pyogenes Cas9 recognizes the PAM 5′-NGG. The first 21 nucleotides of a gRNA bind to the target site via standard Watson–Crick–Franklin base pairing. Upon binding, Cas9 generates a DSB 3 nt upstream of the PAM. (Bottom) DSBs are repaired by one of two general DNA-repair mechanisms: nonhomologous end-joining (NHEJ) and homology-directed repair (HDR). Both pathways are active in germ cells, and the experimenter has little control over which pathway the cell will use to repair the DSB. The NHEJ pathway (left) rejoins the broken ends through an error-prone mechanism that often results in the random insertion and deletion of DNA to generate mutations called indels that can be strategically targeted to disrupt genes and other sequences of interest. The HDR pathway (right) relies on templated repair of the break, which enables precise edits or exogenous DNA to be introduced. During HDR, DNA polymerase uses a template to synthesize DNA that will ultimately be incorporated into the genome. In cells, the template for repair is normally a homologous allele. In gene editing, the researcher supplies a “donor” template that contains desired edits flanked by sequences homologous to the two sides of the DSB (homology arms) that can be recognized by broken genomic DNA to initiate HDR.










