Table 3.
List of PDB structures selected for the comparison of synthetic and natural Abs
| Synthetic Abs | 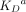 |
Natural Abs | Framework | ||
|---|---|---|---|---|---|
| PDB | Antigen | nM | nM | Antigen | HC/LC |
| 1TZH | Human vascular endothelial growth factor (VEGF) | 1.8 | 4YDK | HIV-1 envelope CD4 binding site | IGHV3/IGKV1 |
| 2QR0 | 7.8 | 6MTO | HIV-1 envelope membrane-proximal external region (MPER) | IGHV1/IGKV3 | |
| 2FJG | 1.5 | 4YWG | HIV-1 envelope V1–V2 region | IGHV4/IGKV3 | |
| 2FJH | 12 | 7LM8 (2)b | SARS-CoV-2 spike glycoprotein RBD | IGHV1/IGKV1 IGHV5/IGKV1 |
|
| 3BDY | 26 | 5GMQ | MERS-CoV spike glycoprotein RBD | IGHV1/IGKV3 | |
| 3PNW | Tudor domain containing protein 3 (TDR3) Tudor domain | 1 | 7KQG | Influenza B hemagglutinin (HA) | IGHV3/IGKV1 |
| 4ZFG | Angiopoietin-2 (ANG2) | 5/5 | 4HG4 | Influenza A hemagglutinin (HA) globular domain | IGHV1/IGKV1 |
| 7RTH | α-Lysozyme nanobody | 21 | 5IBL | IGHV5/IGKV3 | |
| 3R1G | β-Secretase 1 (BACE1) | 1.3 | 3SDY | Influenza A hemagglutinin (HA) stem | IGHV1/IGKV3 |
| 4JQI | β-Arrestin 1 (ARRB1) | NR | 6PZF | Influenza A neuraminidase (NA) | IGHV3/IGKV1 |
| 3G6J | Complement component C3b | 1.2 | 6Q20 | IGHV3/IGKV1 | |
| 1ZA3 | Death receptor 5 (DR5) | 34 | 6WO4 | Hepatitis C envelope glycoprotein E2 | IGHV1/IGKV3 |
| 2H9G | 2 | 6WO5 | IGHV1/IGKV2 | ||
| 3P0Y | Epidermal growth factor receptor (EGFR) | 1.9/0.4 | 5VIG | Zika virus envelope protein DIII Zika virus envelope protein DIII |
IGHV3/IGKV1 |
| 3GRW | Fibroblast growth factor receptor 3 (FGFR3) | <1 | 6DFI | IGHV4/IGKV3 | |
| 6O39 | Frizzled class receptor 5 (FZD5) | 1.7/0.3 | 5VIC | Dengue 1 virus envelope protein DIII | IGHV3/IGKV1 |
| 6O3A | Frizzled class receptor (FZD7) | 41/5.2 | 6OE4 | RSV virus prefusion F0 glycoprotein | IGHV4/IGKV1 |
| 6O3B | 57/4.5 | 6APB | RSV virus postfusion F0 glycoprotein | IGHV2/IGKV1 | |
| 2HFG | BLyS receptor 3 (BR3) | 0.6 | 6BLI | RSV virus G peptide | IGHV4/IGKV1 |
| 3N85 | Epidermal growth factor receptor (HER2) | <1 | 6UVO | IGHV4/IGKV3 | |
| 3K2U | Hepatocyte growth factor activator (HGFA) | <1 | 6N8D | GII.4 2002 norovirus capsid P domain | IGHV3/IGKV3 |
| 4IOF | Insulin-degrading enzyme (IDE) | 4 | 6N81 | IGHV3/IGKV1 | |
| 4DKE | Interleukin-34 (IL-34) | 0.1c | 7L7R (2)b | Crimean–Congo hemorrhagic fever CCHFV Gc prefusion monomer | IGHV3/IGKV1 |
| IGHV4/IGKV1 | |||||
| 4DKF | NR | 3CSY | Ebola glycoprotein | IGHV3/IGKV4 | |
| 5BO1 | Jagged canonical Notch ligand 1 (JAG1) | 0.8 | 6oz9 | IGHV4/IGKV3 | |
| 7KEO | K29-linked di-ubiquitin (Ub) | 3.1 | 7S8H (2)b | Lassa virus glycoprotein 2 | IGHV3/IGKV3 |
| IGHV4/IGKV3 | |||||
| 3DVG | K63-linked di-ubiquitin (Ub) | 8.7 | 6B9J | Vaccinia virus D8 protein | IGHV3/IGKV3 |
| 4K94 | Tyrosine protein kinase KIT domain D4D5 | 0.6 | 7LSE | Tick-borne encephalitis virus (TBEV) envelope domain III (EDIII) | IGHV3/IGKV3 |
| 3SOB | Lipoprotein receptor-related protein 6 (LRP6) | NR | 5VOB | Human cytomegalovirus (HCMV) pentamer | IGHV3/IGKV3 |
| 2QQK | Neuropilin-2 (NRP2) | 0.2 | 5Y11 | Severe fever with thrombocytopenia syndrome virus (SFTSV) glycoprotein N | IGHV5/IGKV3 |
| 3L95 | Notch 1-negative regulatory region (NNR) | 2.5 | 5O14 | Factor H binding protein fHbp (vaccine against Men B) | IGHV5/IGKV1 |
| 4I18 | Prolactin receptor (PRL) | 5.6 | 6XZW | IGHV3/IGKV3 | |
| 6CW2 | Saccharomyces cerevisiae Ada2/Gcn5 | NR | 5O1R | Neisserial heparin binding antigen NHBA (4CMenB vaccine) | IGHV4/IGKV1 |
| 6CW3 | NR | 5D1Q | Staphylococcus aureus IsdB NEAT2 domain | IGHV1/IGKV1 | |
| 5EII | S. cerevisiae anti-silencing factor 1 (ASF1) | NR | 5D1Z | IGHV4/IGKV1 | |
| 5UCB | NR | 4IDJ | S. aureus a-hemolysin | IGHV1/IGKV3 | |
| 5UEA | NR | 7CE2 | Tetanus toxin (TeNT) | IGHV4/IGKV3 | |
| 4XTR | S. cerevisiae Get3-Pep12 | <1 | 6PHB | Plasmodium falciparum vaccine Pfs25 | IGHV4/IGKV1 |
| 3PGF | Escherichia coli maltose binding protein (MBP) | 5/67 | 6PHC | IGHV3/IGKV2 | |
| 5BJZ | 1.5/20 | 7RXP | Plasmodium falciparum C-terminal domain of CSP (ctCSP) | IGHV3/IGKV2 | |
| 5BK1 | 1/152 | 6WOZ | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) | IGHV3/IGKV2 | |
| 5BK2 | 0.9 | 6WQO | IGHV4/IGKV1 | ||
| 5C8J | E. coli YidC | <1 | 6R2S | Plasmodium vivax Duffy binding protein (PvDBP) | IGHV4/IGKV1 |
| 7MDJ | Streptomyces lividans KcsA | 3.7 | 6BFQ | Granulocyte–macrophage colony-stimulating factor (GM-CSF) | IGHV4/IGKV1 |
| 4NNP | S. aureus MntC | 114/189 | 6RLO | CD9 large extracellular loop | IGHV3/IGKV4 |
| 6CBV | E. coli BRIL | 0.3 | 1IQD | Human factor VIII | IGHV1/IGKV3 |
| 7KLH | SARS-CoV-2 spike glycoprotein RBD | <1 | 5XMH | Human IgG1 Fc | IGHV1/IGKV3 |
| 5EU7 | HIV-1 Integrase | NR | |||
| 5W2B | Ebola nucleoprotein | 4.1 | |||
| 6XMI | Salmonella phage 22 terminase | 72 | |||
-
(Abs) Antibodies.
-
aLiterature KD values are provided for synthetic Abs to their target antigen where available. In most cases, KD values were not reported for natural Abs to their target antigen and thus are omitted.
-
b“(2)” indicates that two different Abs within the same PDB file were analyzed.
-
cRefers to the affinity matured Fab variant 1.1.










