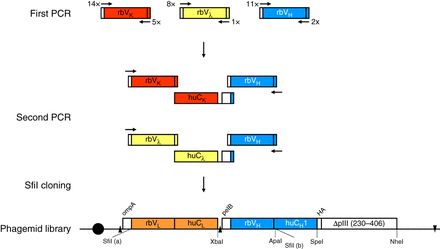
Assembly of chimeric rabbit/human “fragment antigen binding” (Fab) libraries in phagemid pC3C. Shown is a schematic of the workflow for the generation of a phagemid pC3C-based chimeric rabbit/human (rb/hu) Fab library, starting with a first PCR step in which rbVκ-, rbVλ-, and rbVH-encoding cDNAs are amplified from reverse-transcribed mRNA of a naive or immune rabbit antibody repertoire. Diverse primer pairs (14 × 5 for rbVκ, 8 × 1 for rbVλ, and 11 × 2 for rbVH) are used in 100 separate PCRs for each individual rabbit antibody repertoire. Subsequently, a second PCR step fuses the rbVκ and rbVH pools and the rbVλ and rbVH pools via a separately amplified invariable cDNA fragment that comprises huCκ and huCλ, respectively; a downstream ribosome binding site; and pelB. The flanking primers in the second PCR step also introduce the asymmetric SfiI (a) (5′-GGCCC_AGG^CGGCC-3′) and SfiI (b) (5′-GGCC_CCG^TCGGCC-3′) restriction sites, which are cut and ligated into the correspondingly cut phagemid pC3C. Electroporation of the ligation mixture into Escherichia coli completes the phagemid library, which typically comprises 108–1010 independent transformants. Addition of helper phage to the host bacterial cells converts the phagemid-encoding Fab library to a phage-encoding and displaying Fab library. In pC3C, a lac promoter (black circle) drives bicistronic ompA-VL-CL and pelB-VH-CH1-HA-ΔpIII expression cassettes, with ompA and pelB serving as signal peptides, HA (hemagglutinin) as a detection tag, and ΔpIII, which is a C-terminal segment of the minor coat protein pIII of filamentous phage. Shine–Dalgarno sequences are indicated as black triangles, and a transcriptional terminator sequence is depicted as a reverse black triangle. Annotated features of pC3C are discussed in detail elsewhere (see Overview: The pComb3 Phagemid Family of Phage Display Vectors [Rader 2024]).










