Detection of Autophagy in Caenorhabditis elegans
- 1Department of Biology, Queens College-CUNY, Flushing, New York 11367;
- 2The Graduate Center, The City University of New York, New York 10016
Abstract
Autophagy is a dynamic and catabolic process that results in the breakdown and recycling of cellular components through the autophagosomal–lysosomal pathway. Many autophagy genes identified in yeasts and mammals have orthologs in the nematode Caenorhabditis elegans. In recent years, gene inactivation by RNA interference (RNAi) and chromosomal mutations has been useful to probe the functions of autophagy in C. elegans, and a role for autophagy has been shown to contribute to multiple processes, such as the adaptation to stress, longevity, cell death, cell growth control, clearance of aggregation-prone proteins, degradation of P granules during embryogenesis, and apoptotic cell clearance. Here, we discuss some of these roles and describe methods that can be used to study autophagy in C. elegans. Specifically, we summarize how to visualize autophagy in embryos, larva, or adults, how to detect the lipidation of the ubiquitin-like modifier LGG-1 by western blot, and how to inactivate autophagy genes by RNAi.
AUTOPHAGY IN C. ELEGANS
Autophagy is a lysosome-mediated pathway resulting in the degradation and recycling of long-lived proteins, protein aggregates, as well as damaged and old organelles (Levine and Klionsky 2004). It is highly conserved and has been shown to be a fundamental catabolic process in eukaryotes that is required for key developmental and pathological events. Autophagy was first described in mammals, through morphological studies of rat liver cells (Deter et al. 1967). However, it was in the budding yeast Saccharomyces cerevisiae where many autophagy genes (ATG genes) were discovered by screening for mutations that decreased the survival of yeast cells under starvation, as well as mutations that disrupted the cytoplasm-to-vacuole targeting (Cvt) process (Tsukada and Ohsumi 1993; Thumm et al. 1994; Harding et al. 1995, 1996; Hutchins and Klionsky 2001; Klionsky et al. 2003).
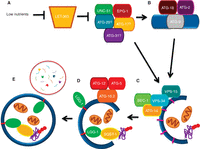
The process of autophagy comprises several distinct steps: formation of a phagophore (also referred to as an isolation membrane or preautophagosomal structure); elongation and closure of the phagophore to form the double-membrane autophagosome; transport and fusion of the autophagosome with a lysosome; and finally degradation of the autophagosomal contents and recycling of degraded material (Fig. 1) (Mizushima 2007; Xie and Klionsky 2007; Nakatogawa et al. 2009). In addition to fusing with a lysosome, an autophagosome can also fuse with an endosome to form a hybrid organelle called the amphisome (Liou et al. 1997; Jing and Tang 1999). When an amphisome or autophagosome fuses with a lysosome, it is referred to as an autophagolysosome (or an autolysosome).
Autophagy in the nematode Caenorhabditis elegans. (A) The process of autophagy has been delineated by studies in yeast and mammalian cells. It is presumed that induction of autophagy begins with the activation of UNC-51, via loss of signaling from the C. elegans ortholog of the target of rapamycin (TOR), LET-363. (B) Autophagosome formation requires the integral protein ATG-9, thought to contribute membrane to the developing autophagosome. (C) Nucleation requires the class III phosphoinositide 3-kinase (PI3K) complex (BEC-1–VPS-34–ATG-14), which recruits downstream autophagy proteins to the isolation membranes (IMs) in mammals or preautophagosomal structure (PAS) in yeast, through the production of phosphatidylinositol 3-phosphate (PI3P; pale purple). (D) Two conjugation complexes (LGG-1 and ATG-12) are required for elongation of the IMs and completion of the developing autophagosome. LGG-1 conjugated to phosphatidylethanolamine (PE, red) binds to both the inner and outer membranes of the autophagosome. LGG-1 also has the ability to bind to autophagic adaptor proteins, such as SQST-1 (encoded by T12G3.1), which bind to polyubiquitylated aggregates. (E) The complete autophagosome eventually fuses with the lysosome (depicted in a red circle), leading to the degradation of cargo within the autophagosome.
The evolutionary conservation of autophagy genes between budding yeast and Caenorhabditis elegans allowed for the identification of genes that encode core components of the autophagic machinery in C. elegans on the basis of genomic sequence homology (Table 1) (Meléndez et al. 2003; Meléndez and Levine 2009). Genetic screens for mutations that disrupt the degradation of P granules led to the discovery of autophagy genes not previously identified in C. elegans on the basis of sequence homology, including: epg-1, the ortholog of yeast ATG13, and epg-8, the ortholog of yeast ATG14 (Table 1) (Tian et al. 2009; Yang and Zhang 2011). Although, the similarities between S. cerevisiae, mammals and C. elegans autophagy proteins suggest that the molecular mechanisms of autophagosome formation might be conserved (Fig. 1; Table 1) (Meléndez and Levine 2009), the presence of genes recently identified in C. elegans that do not have a yeast ortholog could indicate that autophagy involves more-complex membrane dynamics in higher eukaryotes. Hence, it is important to uncover further details about the roles of autophagy genes in autophagosome formation and maturation in C. elegans, and uncover further details about the function of these genes in the different settings where autophagy is required.
Autophagy genes in the nematode Caenorhabditis elegans
The role of autophagy genes in the development of C. elegans has emerged from studies using chromosomal mutations or RNA interference (RNAi) against autophagy genes. Chromosomal mutations exist for many of the autophagy genes found in C. elegans, and many RNAi clones are available (Table 1).
AUTOPHAGY IN C. ELEGANS DEVELOPMENT AND AGING
L1 Arrest after Starvation
Autophagy plays a role mediating the developmental changes associated with survival during extracellular and/or intracellular stress, such as starvation (Levine and Klionsky 2004). In the absence of food, L1 larvae undergo a reversible developmental arrest and can survive for 1–2 wk (Johnson et al. 1984). The insulin-like/IGF-1 signaling (IIS) pathway, which comprises the insulin-like/IGF-1 receptor daf-2 and the Forkhead box protein O transcription factor daf-16, is involved in regulating L1 arrest triggered by starvation (Gems et al. 1998; Baugh and Sternberg 2006; Fukuyama et al. 2006). Interestingly, reduced levels of autophagy have been shown to greatly decrease the survival of starved L1 larvae, emphasizing the importance of autophagy during early stages of development (Kang et al. 2007; Tian et al. 2009, 2010; Alberti et al. 2010; Lu et al. 2011; Yang and Zhang 2011).
Dauer Development
During the first larval stages, animals that are exposed to a limited food supply develop into an alternative L3 larval stage, termed dauer (Albert et al. 1981). Dauer development is associated with morphological and behavioral changes that allow for survival under harsh conditions and stress (Cassada and Russell 1975; Golden and Riddle 1984). The regulation of dauer development has been well characterized and requires the insulin-like/IGF-1, guanylyl cyclase, and transforming growth factor-β signaling pathways, as mutations in any of these pathways can result in a dauer-constitutive phenotype (Daf-c) or a dauer-defective phenotype (Daf-d) (Estevez et al. 1993; Thomas et al. 1993; Gottlieb and Ruvkun 1994; Ren et al. 1996; Schackwitz et al. 1996; Patterson et al. 1997; Birnby et al. 2000; Inoue and Thomas 2000; da Graca et al. 2004). Dauer development is associated with an increase in autophagy, which appears to be required for the cell remodeling associated with proper dauer formation (Meléndez et al. 2003).
Longevity Pathways
In C. elegans, aging is controlled by multiple longevity pathways, such as insulin-like growth factor signaling, target of rapamycin (TOR) signaling, dietary restriction, mitochondrial activity, and germline signaling (Antebi 2007). Recent genetic studies suggest that autophagy interacts with many of these longevity signals to regulate C. elegans aging (Meléndez et al. 2003; Hansen et al. 2008; Toth et al. 2008; Lapierre et al. 2011). Mutants in the insulin-like/IGF-1 receptor (IIR) gene, daf-2, display an increase in autophagy, as detected by an increase in the number of punctate structures labeled with the autophagy marker, GFP::LGG-1, in hypodermal seam cells, a cell type commonly used to visualize autophagy in C. elegans (Meléndez et al. 2003; Hansen et al. 2008). A reduction in autophagy during development, or only during adulthood, shortens the long life span of daf-2 mutants (Meléndez et al. 2003; Hars et al. 2007; Hansen et al. 2008). Reduced food intake without malnutrition, otherwise referred to as dietary restriction, occurs in eat-2 mutants (Avery 1993). These animals lack a nicotinic acetylcholine receptor specific to the pharynx, thereby showing reduced pharyngeal pumping, and have a phenotype of extended life span (Raizen et al. 1995; Lakowski and Hekimi 1998). Consistent with a role for TOR in dietary restriction, eat-2 mutants have reduced TOR signaling, display an increase in autophagy, and require autophagy for their phenotype of long life span (Jia and Levine 2007; Hansen et al. 2008; Toth et al. 2008). The reduction in mitochondrial respiration in isp-1 mutants extends life span (Dillin et al. 2002; Lee et al. 2003), and this phenotype is also dependent on autophagy (Toth et al. 2008). Finally, glp-1/Notch germline-less mutants induce autophagy, and require autophagy for life span extension (Lapierre et al. 2011). Interestingly, protein HLH-30, the ortholog of the mammalian TFEB transcription factor, is required for the life span extension associated with the longevity pathways described above and also regulates autophagy (Lapierre et al. 2013). In conclusion, autophagy is required as part of most longevity pathways in C. elegans, the only exception thus far being the longevity associated with a reduction in protein translation (Pan et al. 2007; Hansen et al. 2008).
Degradation of Paternal Mitochondria
Directly after fertilization, autophagy is induced, resulting in the elimination of spermatozoon-specific organelles, including paternal mitochondria (Al Rawi et al. 2011; Sato and Sato 2011). Whether autophagy also acts in higher eukaryotes to degrade paternal mitochondria is not known; however, an increase in ubiquitylation and the localization of the Atg8-family protein LC3 (Map1lc3a) near the sperm mid-piece at the point of entry are suggestive that this is the case in fertilized mouse zygotes (Al Rawi et al. 2011).
Autophagy in Apoptosis, Necrosis, and Cell Clearance
Although autophagy has a role in homeostasis as an important prosurvival mechanism in response to stress, an excess in autophagy can result in cell death (Kang et al. 2007). Autophagy is also required for necrotic cell death, a type of cell death characterized by the loss of integrity of the cell plasma membrane (Toth et al. 2007; Samara et al. 2008). Additionally, similar to the situation in mammalian cells, BEC-1, a component of the class III phosphoinositide 3-kinase complex (Fig. 1), interacts with an antiapoptotic ortholog of Bcl-2, CED-9, suggesting the existence of cross talk between autophagy and apoptosis (Takacs-Vellai et al. 2005; Erdelyi et al. 2011). Autophagy proteins have been shown to play a role in the proper degradation of apoptotic cell corpses in C. elegans as, in autophagy-deficient animals, apoptotic cells are internalized but not properly degraded (Ruck et al. 2011; Li et al. 2012). Interestingly, rescue experiments indicate that autophagy genes are required within the engulfing cell to promote apoptotic cell degradation (Li et al. 2012).
DETECTING AUTOPHAGY IN C. ELEGANS
Autophagy can be monitored by transmission electron microscopy, fluorescent image analysis of the GFP::LGG-1 reporter or other autophagy reporters (Table 2), and by western blotting. The latter technique takes advantage of changes in the level of lipidation of LGG-1 that occur during autophagy and result in the protein running faster on a gel in its lipidated form. It should be noted that an increase in the number of autophagosomes does not necessarily reflect an induction of autophagy (Klionsky 2012), and it is therefore important to distinguish between induction of autophagy, an increase in autophagic flux, and the accumulation of autophagosomes due to inefficient or blocked autophagy (Klionsky 2012). Usually, it is useful to infer the turnover of autophagosomes in the presence and absence of lysosomal degradation. In C. elegans, this can be achieved by RNAi knockdown of genes with lysosomal function, such as cup-5 (Kostich et al. 2000; Fares and Greenwald 2001; Sun et al. 2011), or the addition of inhibitors such as bafilomycin A1, or chloroquine, routinely used in mammalian cells to inhibit fusion between autophagosomes and lysosomes, which have also been successful in C. elegans (Oka and Futai 2000; Ji et al. 2006; Pivtoraiko et al. 2010). Clearly, the use of multiple assays to verify an increase in functional autophagy is recommended. A comprehensive list of guidelines was recently reported (Klionsky 2012). Here, we describe four protocols for the basic study of autophagy in C. elegans: first, we present a method for the detection of autophagy by using LGG-1 tagged with the fluorescent marker GFP (see Protocol: Detection of Autophagy in Caenorhabditis elegans Using GFP::LGG-1 as an Autophagy Marker [Palmisano and Meléndez 2015a]); second, we give a methodology for monitoring the degradation of P granules, which is dependent on autophagy in embryos (see Protocol: Detection of Autophagy in Caenorhabditis elegans Embryos Using Markers of P Granule Degradation [Palmisano and Meléndez 2015b]); third, we show how western blotting can be used to evaluate the lipidation of LGG-1 and discuss how caution has to be applied to the interpretation of the results of such an analysis (see Protocol: Detection of Autophagy in Caenorhabditis elegans by Western Blotting Analysis of LGG-1 [Palmisano and Meléndez 2015c]); and finally, we present a protocol for investigating the function of autophagy genes by RNAi, a technique that has the advantage of circumventing some of the drawbacks associated with strong loss-of-function mutations (see Protocol: RNAi-Mediated Inactivation of Autophagy Genes in Caenorhabditis elegans [Palmisano and Meléndez 2015d]).
Fluorescent reporters for monitoring autophagy in the nematode Caenorhabditis elegans
ACKNOWLEDGMENTS
We thank all members of the Meléndez laboratory for helpful discussions and comments on the manuscript. We extend special thanks to Melissa Silvestrini and Kristina Ames for comments on an earlier draft. The work in the Meléndez laboratory is supported by a National Science Foundation Research Initiation Award (0818802) and a National Institutes of Health 1 R15 GM102846-01 award; A.M. is an Ellison Medical Foundation New Scholar in Aging and N.J.P. is supported by a CUNY Graduate Center Dissertation award.
- © 2016 Cold Spring Harbor Laboratory Press