Working with Bacteria, Phage, and Plasmids
- 1Université Paris-Saclay, CEA, CNRS, Institut de Biologie Intégrative de la Cellule (I2BC), 91190 Gif-sur-Yvette, France
- 2Departamento de Genética, Facultad de Biología, Universidad de Sevilla, 41080 Sevilla, Spain
- ↵3Correspondence: lionello.bossi{at}i2bc.paris-saclay.fr
Abstract
Methods for the in vivo manipulation of bacterial genomes have improved greatly in recent years because of the discovery of new mechanisms and the gigantic leap forward in DNA-sequencing technology. Many cutting-edge approaches still rely on a variety of technical routines, the correct implementation of which is critical for the success of an experiment. Here, we introduce some of these procedures as used for Escherichia coli and Salmonella enterica. We begin by reviewing the aspects of the biology of these two species that are most relevant for their manipulation in the laboratory.
BACTERIA
Bacteria are unicellular organisms that replicate by binary fission. They comprise the larger domain of the kingdom of prokaryotes, a term rooted in the Greek language (pro, prior; karion, kernel) that denotes the absence of a nuclear structure. Bacterial chromosomal DNA is condensed by proteins in a compact body called the nucleoid, which is in direct contact with the cytoplasm. As a result, the processes of transcription and translation do not take place in separate compartments as they do in eukaryotes; rather they share the same space and are often physically linked and dynamically coupled, meaning that mRNA starts being translated while it is still being synthesized (Johnson et al. 2020; Wang et al. 2020; Webster et al. 2020; York 2020).
Bacteria are broadly divided into two major groups on the basis of their response to the Gram-staining procedure, which probes the chemistry of the cell wall. Gram-positive bacteria have a thick multilayered shield of peptidoglycan surrounding the plasma membrane that is in direct contact with the outside environment. Peptidoglycan is a rigid, net-like polymer composed of long polysaccharide chains held together by peptide bridges (Pazos and Peters 2019). It determines the shape of bacteria and protects them from osmotic challenges and other environmental insults. Peptidoglycan can be cleaved by the enzyme lysozyme, used in most procedures for lysing bacterial cells. Gram-negative bacteria also have a peptidoglycan sacculus; however, it is much thinner than that in Gram-positive bacteria and is itself surrounded by a second lipid bilayer, the outer membrane. The outer membrane plays an important role by acting as a barrier excluding toxic molecules and contributing to the structural integrity of the peptidoglycan and other cell components. Much of the robustness of the outer membrane is due to the presence of a thick layer of lipopolysaccharides (LPSs). LPSs are long molecules with a fatty acid component that anchors to the membrane and a complex polysaccharide moiety, the side chains of which, collectively called the O-antigen, extend beyond the cell surface (Wang and Quinn 2010; Liu et al. 2020). Variability in the sugar composition of the O-antigen accounts for a large part of the diversity between strains of a given species of Gram-negative bacteria. Of note, LPS serve as receptors for the primary binding of bacteriophages including generalized transducing phages P1 of Escherichia coli (Zhang et al. 2020) and P22 of Salmonella enterica (Susskind and Botstein 1978). These two bacteria are the best-characterized members of the Gram-negative group, widely used as models in genetic research for nearly 80 years. The best-studied Gram-positive bacteria include Bacillus subtilis and Staphylococcus aureus.
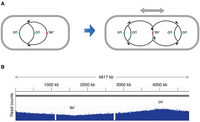
Most bacteria have a single covalently closed circular chromosome, but there are some notable exceptions, such as Vibrio cholerae, the genome of which is organized in two separate circular chromosomes (Escudero and Mazel 2017), and Streptomyces, which have a single linear chromosome (Hopwood 2006). In Escherichia coli and Salmonella, replication of chromosomal DNA starts at a unique origin and proceeds bidirectionally toward a terminus region (Prescott and Kuempel 1972). Because of the 5′ to 3′ polarity of DNA synthesis, the two DNA strands are elongated asymmetrically: One strand, the so-called leading strand, is replicated continuously, whereas the other strand, the lagging strand, is replicated in the form of short DNA fragments, the so-called Okazaki fragments, that are subsequently joined together by DNA ligase. When growing in nutrient-rich medium, E. coli and Salmonella bacteria divide faster than the time needed for the replisome complex to travel from the origin to the terminus. This is made possible by the temporal overlap of consecutive rounds of replication; that is, DNA replication initiates before the previous round is completed (Cooper and Helmstetter 1968). As a result, genes that are close to the origin are present in higher copy number than distal genes (Fig. 1A). In asynchronous culture, this phenomenon manifests as a gene-dosage gradient along the two halves of the chromosome (Fig. 1B). The fact that, under these conditions, the bacterium is essentially merodiploid in part of its chromosome has some implications for genetic manipulations. In particular, if introducing a recessive allele by recombination, the phenotype associated with this allele will not manifest before the sister chromosomes have segregated in daughter cells (Newcombe 1948). In cases in which the resident allele is lethal under the selection conditions, giving sufficient time for the sister chromosomes to segregate is essential to recover live recombinants.
Gene-dosage gradient in bacterial DNA replication. (A) Schematic diagram of the replicating chromosome during cell growth. (B) Read-count profile of randomly fragmented chromosomal DNA from exponentially growing Salmonella in a high-throughput DNA-sequencing experiment. (The two gaps in the profile correspond to the locations of two prophages missing in the strain used here.)
TRANSDUCING PHAGES
Phage-mediated generalized transduction was initially discovered by N. Zinder and J. Lederberg in Salmonella and described in an article published in 1952 (Zinder and Lederberg 1952). Shortly after, similar findings were reported in E. coli (Lennox 1955). The phages involved, P22 in Salmonella and P1 in E. coli, are still widely used for transduction in these organisms today. Although, in many gene-engineering applications, transduction has been replaced by more convenient techniques, it remains the method of choice for transferring newly isolated mutations or gene constructs into a “clean” genetic background.
P1 and P22 are temperate bacteriophages that infect the respective hosts by binding to the LPS core and O-antigen side chain on the outer membrane, respectively (Eriksson and Lindberg 1977; Sandulache et al. 1984). The two phages have markedly different genome sizes, the P1 genome (∼94 kb) being more than twice the length of the P22 genome (∼42 kb). At the latest stages of the infection cycle, the DNA from both phages replicates by a rolling-circle mechanism, producing long concatemers containing multiple copies of the phage genome. These concatemers are packaged into phage heads by a “headful” mechanism: Packaging is initiated at a specific sequence on the DNA called a pac site, and then a phage-encoded nuclease moves down the concatemer, cutting approximately every 110 kb in P1 DNA (Sternberg and Coulby 1987) and every 48 kb in P22 DNA (Casjens and Hayden 1988). Because these lengths are greater than the respective genome lengths, packaged fragments contain repeated sequences at their ends, a feature referred to as terminal redundancy. Homologous recombination between the direct repeats allows circularization of the injected linear fragment upon infection of the bacterial host.
E. coli and Salmonella genomes contain sequences similar to the pac sites of the corresponding phages. When P1 and P22 infect cells, occasionally their encoded nucleases cleave one of these chromosomal sites, resulting in the sequential packaging of chromosomal DNA segments into phage heads. The particles carrying these segments (transducing particles) can inject them into a new host. The DNA can then recombine into the chromosome by homologous recombination. As the phage can transfer DNA fragments from all regions of the chromosome, this process is called generalized transduction (Zinder and Lederberg 1952; Lennox 1955).
PLASMIDS
Plasmids are extrachromosomal DNA elements found in many types of cells, most often, although not always, in the form of covalently closed circles. Although generally dispensable for the life of the host cell, plasmids can profoundly affect its physiology and behavior. In bacteria, plasmids often carry genes that improve bacterial fitness or confer virulence traits or resistance to antibiotics. In contributing to the circulation of these genes in the environment, they strongly impact the fields of bacterial ecology, microbial evolution, and clinical microbiology. In molecular biology, plasmids are intimately linked to the birth of molecular cloning, having provided the backbone of the early cloning vectors. Today plasmids are used in a myriad of gene-editing applications and constitute irreplaceable tools in the experimental inventory of molecular geneticists.
Plasmids replicate independently of the bacterial chromosome but using the replication machinery of the host. Only one plasmid-encoded function, the Rep protein, is most often (although not always) required for this process. Rep binds to a specific site in the plasmid DNA and, often acting in concert with the host protein DnaA, promotes local unwinding of the double helix and assembly of the replication–initiation complex (Ingmer and Cohen 1993). Thus, the Rep-binding site defines the origin (ori) of plasmid replication. Although variable at the DNA-sequence level, replication origins share conserved features—namely, a high content of AT base pairs and the presence of short direct repeats, called iterons, that are the sites where Rep proteins bind (Sugiura et al. 1993). Rep proteins are typically encoded in the proximity of their binding sites. However, they can also function when provided in trans. This property was exploited in the development of the so-called suicide vectors, plasmids from which rep genes have been removed and placed elsewhere so that they can only replicate in those specific backgrounds (Kolter et al. 1978). A typical example of this is pGP704, which carries the minimal replication origin of plasmid R6K (the γ ori), normally dependent on the protein π for replication (Miller and Mekalanos 1988). Plasmid pGP704 will only replicate in strains that harbor the π-encoding pir gene in the chromosome. When introduced into a strain lacking the pir gene, pGP704 derivatives will produce stable transformants only if they carry a DNA insert that allows them to integrate into the chromosome via homologous recombination (Miller and Mekalanos 1988). This strategy has been widely used for the construction of insertion mutants and for the transfer of mutations isolated on a cloned gene into the chromosome by allelic replacement. A further example of conditional replicons is the temperature-sensitive derivatives of plasmid pSC101 (Hashimoto-Gotoh and Sekiguchi 1977). Plasmids carrying mutation repA101ts replicate normally and are stably maintained at 30°C, but they are rapidly lost when the temperature is increased to 37°C—a convenient feature when it is desirable to remove a plasmid after its utilization in a strain. Two repA101ts-based replicons, plasmids pKD46 and pCP20, are currently used in λ Red recombineering experiments.
PROTOCOLS
In the accompanying protocols, we provide step-by-step details that are implemented in the study of bacteria in the laboratory. They include details for handling enteric bacteria and bacteriophages (see Protocol: Basic Bacteriological Routines [Figueroa-Bossi et al. 2022a] and Protocol: Working with Phage P22 [Figueroa-Bossi et al. 2022b]) and extracting plasmid and chromosomal DNA from bacterial cells (see Protocol: Preparing Plasmid DNA from Bacteria [Figueroa-Bossi et al. 2022c] and Protocol: Preparing Bacterial Genomic DNA [Figueroa-Bossi et al. 2022d]). Also included is a method for bacterial transformation (Protocol: Quick Transformation with Plasmid DNA [Figueroa-Bossi et al. 2022e]).
ACKNOWLEDGMENTS
Work in our laboratory was supported by the Centre National de la Recherche Scientifique (CNRS) and by the Agence Nationale de la Recherche (ANR-15-CE11-0024-03), France.
Footnotes
-
From the Experiments in Bacterial Genetics collection, by Lionello Bossi, Andrew Camilli, and Angelika Gründling.