Engineering of Affibody Molecules
- ↵1Correspondence: ssta{at}kth.se
Abstract
Affibody molecules are small, robust, and versatile affinity proteins currently being explored for therapeutic, diagnostic, and biotechnological applications. Surface-exposed residues on the affibody scaffold are randomized to create large affibody libraries from which novel binding specificities to virtually any protein target can be generated using combinatorial protein engineering. Affibody molecules have the potential to complement—or even surpass—current antibody-based technologies, exhibiting multiple desirable properties, such as high stability, affinity, and specificity, efficient tissue penetration, and straightforward modular extension of functional domains. It has been shown in both preclinical and clinical studies that affibody molecules are safe, efficacious, and valuable alternatives to antibodies for specific targeting in the context of in vivo diagnostics and therapy. Here, we provide a general background of affibody molecules, give examples of reported applications, and briefly summarize the methodology for affibody generation.
INTRODUCTION
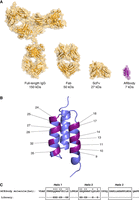
Affibody molecules are small (6- to 7-kDa, 58-amino acid) affinity proteins of a tightly packed three-helical bundle architecture with a hydrophobic core, engineered to bind specific molecular targets (Fig. 1; Ståhl et al. 2017). The scaffold is based on the Z-domain, which originates from the introduction of two point mutations in the immunoglobulin (Ig) G-binding B-domain of staphylococcal protein A. Target specificity is achieved by combinatorial protein engineering from randomization of typically 13 surface-exposed residues on helices 1 and 2, followed by selection or high-throughput screening using technologies such as phage, bacterial, or yeast display. Affibody molecules can be recombinantly produced in bacterial hosts such as Escherichia coli, both as single domains and as more complex fusion constructs, greatly reducing the cost and complexity of manufacturing. They are easily engineered and are amenable for modular extension of functional or binding moieties through simple genetic fusion. The small size of the affibody scaffold provides alternative avenues and distinct advantages that have the potential to circumvent many of the challenges typically associated with full-length antibodies.
Structure of the affibody molecule. (A) Three-dimensional structures, showing a full-length IgG antibody (PDB ID 1HZH), a Fab antibody fragment (PDB ID 6MH2), an scFv antibody fragment (PDB ID 1X9Q), and an affibody molecule (PDB ID 2B88). The proteins are shown in scale, and the molecular weights are indicated below the corresponding protein name. (B) Three-dimensional structure of an affibody molecule, with the 13 randomized positions on helices 1 and 2 indicated. (C) Amino acid sequence in single-letter code of the protein A–derived Zwt domain, with the three α-helices boxed and the randomized positions indicated.
In this review, we provide a brief overview of affibody molecules, discussing various applications, the current state of the affibody technology, and methodologies for affibody generation. First, we discuss examples of biomedical applications that have been investigated using affibody molecules and translational successes thus far in clinical evaluations. We also discuss affibody molecules in the context of radionuclide molecular imaging. Last, we discuss methods by which affibody molecules are generated, looking specifically at approaches for library design and common selection strategies.
BIOMEDICAL APPLICATIONS OF AFFIBODY MOLECULES
The small size of affibody molecules offers great advantages for biomedical applications. For instance, it allows for rapid blood clearance by renal excretion, which offers a favorable pharmacokinetic profile for diagnostic imaging (Tolmachev and Orlova 2020). Additionally, affibodies readily extravasate from blood and efficiently penetrate tissues, offering favorable tumor-targeting properties (Schmidt and Wittrup 2009). Furthermore, affibody molecules may alleviate logistical challenges and improve patient compliancy due to the potential for alternative administration routes such as subcutaneous, pulmonary, oral, and ocular delivery, which is being explored with promising results (Ståhl et al. 2017). For therapeutic applications, which often necessitate longer blood circulation for prolonged drug exposure and reduced dosing frequency, the serum half-life can be readily extended by various technologies. The most well-investigated half-life extension strategy for affibody molecules is genetic fusion to an engineered albumin-binding domain, conferring a biodistribution profile and half-life similar to that of serum albumin (Jonsson et al. 2008).
Research in the field has primarily been focused on medical applications, with more than 650 publications to date describing affibody molecules versus more than 50 molecular targets relevant for oncology, neurodegenerative diseases, and inflammatory disorders (Ståhl et al. 2017). The studies span from engineering efforts and in vitro cell assays to preclinical and clinical evaluations. Several affibody molecules have been engineered to bind medically relevant targets with subnanomolar—and even subpicomolar—affinity, such as platelet-derived growth factor receptor beta (PDGFRβ) (400 pM) (Lindborg et al. 2011), vascular endothelial growth factor receptor 2 (VEGFR2; 200 pM) (Fleetwood et al. 2016), epidermal growth factor receptor (EGFR; 160 pM) (Andersson et al. 2016), human epidermal growth factor receptor 2 (HER2) (22 pM) (Orlova et al. 2006), HER3 (21 pM) (Malm et al. 2013), amyloid β peptide (20 pM) (Lindberg et al. 2015; Boutajangout et al. 2019), and interleukin (IL)-17A (0.3 pM) (Ståhl et al. 2017). These high-affinity binders have been used to explore various treatment strategies, including diagnostic molecular imaging of tumors, receptor signaling inhibition, ligand trapping, and cytotoxic payload delivery. Currently, three affibody molecules have proceeded to different stages of clinical development and testing. A HER2-targeting affibody molecule, ABY-025, labeled with the radioactive compound gallium-68, has been used, together with positron-emission tomography (PET) imaging, to stratify patients with HER2-positive primary and metastatic breast cancer in a phase I clinical study (Velikyan et al. 2019), and patients are currently being recruited for phase II/III. In addition, a fluorescently labeled EGFR-targeting affibody molecule, ABY-029, is being evaluated for guided resection of tumors in patients with recurrent glioma (Sexton et al. 2013). A treatment for psoriatic arthritis is being investigated using a dimeric affibody, ABY-035, to neutralize IL-17A, which has shown a favorable safety profile and efficacy over the standard of care in phase II clinical trials (https://acelyrin.com/press/acelyrin-affibody-ab-inmagene-announce-data-from-global-phase-2-trial-of-izokibep). Indeed, results from clinical studies have thus far shown that affibody molecules are safe and tolerable in humans. ABY-035 has shown rapid and sustained efficacy in patients, as well as favorable safety and tolerability data, and the majority of adverse events have been mild and resolved during treatment.
The small size and robustness of affibody molecules are particularly useful in the context of radionuclide molecular imaging. More than 200 publications concerning preclinical affibody-based tumor imaging are currently available. The most extensively studied cancer biomarkers using affibody-based tracers are the HER family of receptors (Rinne et al. 2021), including EGFR, HER2, and HER3. The fast pharmacokinetics associated with these molecules offers improved contrast and more sensitive scans at earlier time points, usually just a few hours after injection. Affibody molecules generally show high thermal and chemical stability, which facilitates downstream processes such as purification, conjugation, and radionuclide labeling. Their small size also has implications for the design of affibody-based imaging tracers. Minor alterations may considerably affect their overall charge and hydrophilicity, meaning that biodistribution is tunable by optimization of various parameters, such as chelators, nuclides, tags, and amino acid composition, which has the potential to minimize off-target interactions without noticeably affecting affinity for the cancer-associated target antigen. High affinity and, thus, longer tumor retentions, which are usually attainable for affibody molecules, are necessary to achieve high contrast for imaging molecular targets with slow internalization, in particular for the detection of targets with low expression levels (Tolmachev et al. 2012).
AFFIBODY GENERATION AND LIBRARY DESIGN
Affibody libraries are typically constructed by randomizing 13–15 positions on helices 1 and 2. By using a predefined set of codons, a large naive repertoire of tens of billions of affibody variants is created, from which unique binding specificities can be selected (Fig. 1). Degenerate NNK codons (where N = A/C/G/T and K = G/T) have traditionally been used for the construction of affibody libraries. Most libraries today, however, are made using a mixture of trinucleotide codons during DNA library synthesis, providing more controlled diversification, decreased bias, and limited introduction of stop codons (Arunachalam et al. 2012). Cysteine residues are absent in the affibody scaffold and are usually omitted in the library design, to enable post-selection introduction of a solitary cysteine for site-specific thiol-coupling of functionalizing groups, such as metal chelators, fluorophores, biotin, and cytotoxic compounds. Proline is also often excluded due to its helix-breaking propensity. Moreover, the small size of the affibody gene (174 bp) makes synthesis and molecular cloning of DNA-encoded affibody libraries straightforward. The randomized positions are located within a sequence of merely 78 bp on helices 1 and 2 (Fig. 1). The DNA library can thus be synthesized as a single randomized oligonucleotide, that includes, for example, restriction enzyme sites or overlapping sequences for restriction-free gene assembly to the display plasmid of choice, as described, for instance, in Protocol: Cloning of Affibody Libraries for Display Methods (Ståhl et al. 2023).
Affibody molecules generated from naive selections typically have affinities in the nanomolar range. If higher affinities are desired, a maturation library is usually constructed to improve affinity. When designing the maturation library, amino acid frequencies and alanine scanning can be used to elucidate the importance of certain amino acids and positions for the binding interaction. For example, a maturation library was created for a HER3-binding affibody, in which the binding contributions of the 13 residues were weighed based on the results from alanine scanning (Malm et al. 2013). Each of the 13 positions were randomized using an equal mixture of 17 trinucleotide codons, but the original residues were preserved to a degree proportionate to their binding contribution (a method often denoted as “soft mutagenesis”), resulting in binders with affinities in the low picomolar range.
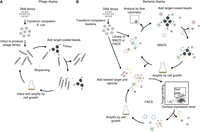
Selections of affibody molecules are performed by panning against the target antigen in a setting appropriate for the choice of display system. Phage display is still the most commonly used method for generation of new affibody molecules, and detailed instructions for construction of phage-displayed affibody libraries and phage-display biopanning using such libraries are described in Protocol: Cloning of Affibody Libraries for Display Methods (Ståhl et al. 2023) and Protocol: Selection of Affibody Molecules Using Phage Display (Hjelm et al. 2023), respectively (Fig. 2A). In addition to phage display, selection of affibody molecules has also been shown using other display methods, such as yeast display (Stern et al. 2019), ribosome display (Grimm et al. 2011), E. coli display (Andersson et al. 2019), and staphylococcal display (Malm et al. 2013). Methods for generating libraries in E. coli and Staphylococcus carnosus, together with details on selection and screening from such libraries are described in Protocol: Selection of Affibody Molecules Using Escherichia coli Display (Dahlsson Leitao et al. 2023) and Protocol: Selection of Affibody Molecules Using Staphylococcal Display (Löfblom et al. 2023). The main advantage of using cell-based display systems is that the selection can be performed using fluorescence-activated cell sorting (FACS) (Löfblom 2011). For FACS, the target antigen is fluorescently labeled and incubated with cells that each display 104–105 copies of a unique affibody variant. During selection, binding populations can be separated from nonbinding populations in real time based on fluorescent signals, allowing enrichment to be observed and followed throughout each selection round. Surface expression of the affibody variants is generally assessed by the inclusion of an albumin-binding domain in the displayed protein construct, followed by incubation with fluorescently labeled serum albumin (Löfblom et al. 2005). For cell-based display systems, the complexity of naive libraries is typically reduced by using magnetic-activated cell sorting in a preenrichment step before library sorting using FACS (Fig. 2B), which is described as part of Protocol: Selection of Affibody Molecules Using Escherichia coli Display (Dahlsson Leitao et al. 2023).
Common selection strategies for generating affibody molecules. Schematic overview of the selection processes in (A) phage display and (B) bacterial display. In phage display, the affibody library is displayed on bacteriophages and panned against the target coated on a solid support, such as paramagnetic beads. The binding population associated with the beads is isolated by washing away the nonbinding population. The binding population is amplified and the procedure is repeated two to four times to enrich for affibody variants that bind to the target. In bacterial cell display, the affibody library is displayed on the surface of bacterial cells and incubated with fluorescently labeled target and reporter molecules, followed by the isolation of binding populations using fluorescence-activated cell sorting (FACS). The binding population is amplified, and the procedure is repeated two to four times to enrich for affibody variants that bind to the target. Note that, for larger libraries, magnetic-activated cell sorting (MACS) is advisable to reduce the library sizes to allow efficient FACS.
CONCLUSIONS
Affibody molecules are an interesting and versatile class of affinity proteins that are being explored for therapeutic and diagnostic applications for several diseases, as well as for other biotechnological applications. The unique characteristics of the affibody molecule have the potential to complement antibody-based therapies and improve on current cancer diagnostic imaging technologies. Generation of novel binding specificities from large affibody libraries against virtually any target is possible and straightforward.
ACKNOWLEDGMENTS
The research on affibodies and their selection systems was funded by the Swedish Agency for Innovation VINNOVA (2019/00104 and the CellNova Center 2017/02105), the Knut and Alice Wallenberg Foundation through the Wallenberg Center for Protein Technology (KAW 2019.0341 and KAW 2021.0197), the Swedish Cancer Society (grants 201090 PjF; 222023 Pj01H), the Swedish Research Council (2019-05115), the Swedish Brain Foundation (grants FO2018-0094, FO2021-0407, FO2022-0253), the Tussilago foundation (FL-0002.025.551-7), the Strategic Research Area Neuroscience (StratNeuro), as well as the Schörling Family Foundation via the Swedish FTD Initiative.
Footnotes
-
From the Advances in Phage Display collection, edited by Gregg J. Silverman, Christoph Rader, and Sachdev S. Sidhu.