Introduction to CRISPR and Its Use in Drosophila
- 1Department of Neuroscience, Brown University, Providence, Rhode Island 02912, USA
- 2Carney Institute for Brain Science, Brown University, Providence, Rhode Island 02912, USA
- 3Pediatrics, Cell & Developmental Biology, University of California, San Diego, La Jolla, California 92093, USA
- 4Biological Sciences, Cell & Developmental Biology, University of California, San Diego, La Jolla, California 92093, USA
- ↵5Correspondence: oconnorgiles{at}brown.edu; jwildonger{at}ucsd.edu
Abstract
The preeminence of Drosophila genetics has led to key discoveries in biology across a variety of fields and disciplines. The advent of CRISPR gene editing has expanded the toolkit of genetic reagents that can be applied to manipulate and observe genes, RNAs, and proteins in an in vivo context. This review describes CRISPR and its use as a transformative gene editing tool in Drosophila. We focus on the canonical pathway in which the Cas9 nuclease is directed to specific sequences by guide RNA (gRNA), where cleavage leads to DNA repair by one of two main cellular pathways: nonhomologous end joining (NHEJ) or homology-directed repair (HDR). The error-prone NHEJ pathway can be appropriated to disrupt targeted sequences, enabling a variety of loss-of-function studies. Induction of the HDR pathway allows precise editing, including defined deletions, the introduction of specific sequence changes, and the incorporation of fluorescent and epitope tags. These approaches have increased the power of Drosophila genetics and been successfully used to conduct in vivo structure–function studies, study disease-associated variants, and follow protein dynamics.
INTRODUCTION
CRISPR, which stands for clustered regularly interspaced palindromic repeats, is a bacterial immune system that has been adopted as a straightforward method for editing the genomes of a wide variety of organisms, including numerous Drosophila species (Bier et al. 2018). In gene editing, CRISPR is used to induce DNA breaks in precise genomic locations. As cells must repair DNA breaks to survive, this provides a window of opportunity for editing the genome at targeted sites during the repair process. Jinek et al. (2012) showed that a simplified CRISPR system, adopted from Streptococcus pyogenes, could be programmed to cleave DNA at defined sites in vitro. By 2013, multiple groups showed its utility for gene editing in mammalian cells, and the system was quickly adopted to generate heritable mutations in flies (Bassett et al. 2013; Cong et al. 2013; Gratz et al. 2013; Jinek et al. 2013; Mali et al. 2013; Yu et al. 2013). This rapid time line demonstrates the profound usefulness and relative simplicity of CRISPR and reflects the extensive bodies of work in CRISPR biology, DNA repair, and gene editing that informed its development. CRISPR has now become the primary tool for generating a wide variety of function-probing genetic reagents in flies. For Drosophila neurobiologists, CRISPR has become an indispensable tool for generating mutations including disease-associated variants, conducting in vivo structure–function studies, tagging endogenous proteins to follow their dynamic localization, and much more.
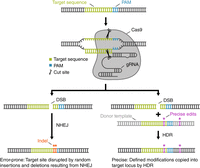
There are three critical components to CRISPR gene-editing systems: (1) Cas9, the enzyme that generates double-stranded DNA breaks (DSBs) through its two nuclease domains, (2) a synthetic RNA, called a guide RNA (gRNA), that can be readily programmed to bind specifically to target sequences in the genome, and (3) a motif adjacent to the genomic site bound by the gRNA called a protospacer-adjacent motif, or PAM, which Cas9 requires to cleave double-stranded DNA (Fig. 1). The gRNA binds Cas9 and recruits the enzyme to the target sequence in the genome. Because recognition between a gRNA and its genomic target site occurs through simple Watson–Crick–Franklin base pairing, the DNA-recognition sequence of the gRNA can be tailored to bring Cas9 to virtually any site in the genome. If the genomic target site is flanked by the appropriate PAM sequence, Cas9 will generate DSBs that are subsequently repaired by the cell.
Gene editing with CRISPR–Cas9: components and DNA-repair pathways. The CRISPR–Cas9 gene-editing system uses two simple components to induce targeted double-stranded breaks (DSBs), the repair of which provides an opportunity to edit the genome. (Top) The Cas9 enzyme cleaves both strands of DNA using its two nuclease domains and a guide RNA (gRNA). As its name implies, the gRNA “guides” Cas9 to its target site, which must be adjacent to a 3′ protospacer-adjacent motif (PAM) for cleavage to occur. The widely used Streptococcus pyogenes Cas9 recognizes the PAM 5′-NGG. The first 21 nucleotides of a gRNA bind to the target site via standard Watson–Crick–Franklin base pairing. Upon binding, Cas9 generates a DSB 3 nt upstream of the PAM. (Bottom) DSBs are repaired by one of two general DNA-repair mechanisms: nonhomologous end-joining (NHEJ) and homology-directed repair (HDR). Both pathways are active in germ cells, and the experimenter has little control over which pathway the cell will use to repair the DSB. The NHEJ pathway (left) rejoins the broken ends through an error-prone mechanism that often results in the random insertion and deletion of DNA to generate mutations called indels that can be strategically targeted to disrupt genes and other sequences of interest. The HDR pathway (right) relies on templated repair of the break, which enables precise edits or exogenous DNA to be introduced. During HDR, DNA polymerase uses a template to synthesize DNA that will ultimately be incorporated into the genome. In cells, the template for repair is normally a homologous allele. In gene editing, the researcher supplies a “donor” template that contains desired edits flanked by sequences homologous to the two sides of the DSB (homology arms) that can be recognized by broken genomic DNA to initiate HDR.
Cells rely on two broad categories of DNA-repair mechanisms (Fig. 1). The first mechanism, nonhomologous end-joining (NHEJ), is active in many cell types, including germ cell progenitors, and it is the predominant pathway used in postmitotic neurons. In NHEJ, cells repair DNA breaks through ligation of broken ends. This process is imprecise and often accompanied by the gain or loss of DNA at the repair site. The resulting insertions and deletions are commonly referred to as indels. These tend to be small, on the order of a few base pairs, but larger indels are also observed. The second mechanism is homology-directed repair (HDR). HDR is a precise process involving templated repair across the break that can occur through a number of distinct pathways. HDR involves broken DNA ends recognizing and binding to complementary homologous sequence (normally on a sister chromatid or homologous chromosome), which serves as a template for the synthesis of DNA to precisely repair the break.
The generation of targeted DSBs using CRISPR creates an opportunity for researchers to co-opt the cell's repair processes to edit genomic DNA. Researchers working with model organisms use both NHEJ and HDR for gene editing. Creating targeted indels via the error-prone NHEJ pathway is an efficient mechanism for generating loss-of-function alleles or disrupting other genomic sequences. In contrast, by supplying exogenous repair templates, referred to as donors, the HDR pathway can be used to introduce mutations, generate precise deletions, or introduce exogenous sequences such as those encoding fluorescent and peptide tags. Donor templates must include sequences that are homologous to both sides of the break site, called homology arms, as well as any new sequence to be incorporated. It is critical to note that the experimenter has little control over whether the cell employs the NHEJ or HDR pathway. Thus, the keys to successful gene editing are supplying the necessary reagents for the preferred pathway and a solid strategy for selecting the desired repair events.
Researchers have also extended the use of CRISPR to a broad array of applications of interest to Drosophila neurobiologists, including cell-specific editing; prime editing, in which only one strand of DNA is nicked and the desired edits are coded in extended gRNAs called prime editing gRNAs (pegRNAs); RNA targeting with Cas13; and dCas9 approaches in which nuclease-dead Cas9 is used to shuttle other enzymes to precise locations in the genome (Lin et al. 2015; Ghosh et al. 2016; Buchman et al. 2020; Huynh et al. 2020; Port et al. 2020; Zirin et al. 2020; Bosch et al. 2021). For more information on these approaches, we encourage the reader to explore the comprehensive links at the FlyBase CRISPR Wiki page: wiki.flybase.org/wiki/FlyBase:CRISPR.
In our associated protocol, we introduce a step-by-step procedure for the most commonly used CRISPR strategies for making heritable mutations in flies: (1) NHEJ to create small random indels that can lead to frameshifting mutations or larger deletions and (2) HDR to make specific sequence changes or insert exogenous sequences such as molecular tags (see Protocol: Generating CRISPR Alleles in Drosophila [Gratz et al. 2023]). Note that this approach can be applied in any species with modifications for reagent delivery.
Footnotes
-
From the Drosophila Neurobiology collection, edited by Bing Zhang, Ellie Heckscher, Alex C. Keene, and Scott Waddell.