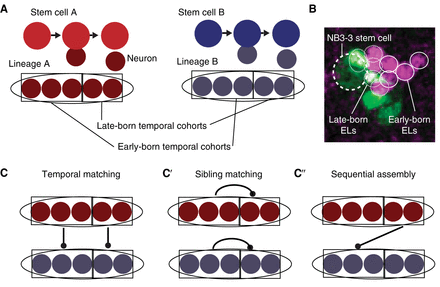
Stem cell lineages are organized into temporal cohorts in Drosophila. (A) In this illustration, stem cell A (red) and stem cell B (blue) generate two distinct lineages (ovals) of neurons. Both lineages are subdivided into temporal cohorts (boxes). Each circle represents a cell, with arrows representing cell division. (B) An image shows early-born and late-born Even-skipped (Eve)-expressing lateral (EL) neurons (small circles, each ∼3 µm in diameter) from the NB3-3 stem cell (large dashed circle at left). This late-stage embryo was stained with an anti-Even-skipped antibody (magenta) to label all EL neurons (small circles) and costained with anti-GFP (green). In this embryo, GFP expression is being driven by an enhancer, “11F02” (genotype: 11F02-GAL4 > UAS-nls-GFP). (C) Lineages can be connected to each other in different patterns. Symbols are as in A and, additionally, the line with a dot on the end represents synapses between neurons of one temporal cohort and another temporal cohort. In temporal matching, early-born neurons and late-born neurons from different lineages synapse with each other. (C′) In sibling matching, neurons from the same lineage synapse with each other. (C″) In sequential assembly, neurons from a late-born temporal cohort in one lineage synapse with neurons from an early-born temporal cohort in a second lineage. Because neurons make multiple connections, these models are not mutually exclusive.










