The Study of Sensorimotor Circuit Assembly in Drosophila melanogaster Embryos and Larvae
- 1Department of Molecular Genetics and Cell Biology, The University of Chicago, Chicago, Illinois 60637, USA
- 2Institute for Neuroscience, The University of Chicago, Chicago, Illinois 60637, USA
- 3Committee on Neurobiology, The University of Chicago, Chicago, Illinois 60637, USA
- 4Committee for Development, Regeneration, and Stem Cell Biology, The University of Chicago, Chicago, Illinois 60637, USA
- ↵5Correspondence: heckscher{at}uchicago.edu
Abstract
In animals, movement is generated by the activity of motor circuits housed in the vertebrate spinal cord or the arthropod nerve cord. How motor circuits form is a fundamental question, with wide-ranging impacts on the fields of development, neurobiology, medicine, evolution, and beyond. Until recently, studying circuit assembly had been experimentally difficult, with a paucity of suitable models. Due to the introduction of novel neuroscience tools (calcium imaging, optogenetics, connectomics), Drosophila embryos and larvae can be used as models to study motor circuit assembly. Here, we briefly review the knowledge relevant to motor circuit assembly in Drosophila larvae. We discuss the larval body and its movements, larval neurons and circuits in the motor system, and how the generation of neural diversity starting from stem cells relates to circuit formation. The long-term goal of Drosophila research in this field is to identify developmental rules, determine when the rules apply, generate an integrated understanding of motor circuit development, and uncover molecular mechanisms driving the assembly process. Motor circuits are an ancient part of the nervous system, and so far, the developmental programs guiding motor circuit assembly appear to be largely conserved across phyla. Thus, as methods improve in other systems, findings in Drosophila will provide foundational concepts that will inspire hypotheses in those systems.
INTRODUCTION
Movement must be precise and robust, tuned specifically to the environment, and yet, the most complex movements often appear effortless. In animals, the nervous system generates movements using the activity of motor circuits that are housed in the arthropod nerve cord or the vertebrate spinal cord. Because motor circuits are the only circuits to contain motor neurons, they are also the only circuits that can directly generate movement. In addition to generating movement, motor circuits also process somatosensory stimuli in parallel (e.g., pain, touch, vibration, temperature, and self-movement). Motor circuits are composed of similar but nonidentical segments arrayed along the rostrocaudal (anterior–posterior) axis.
In most organisms, the peripheral parts of the motor system (sensory neurons, motor neurons, and muscles) are far better understood than central circuits. Below, we review some of the general principles underlying motor circuits. Fundamentally, motor circuits are layered structures, with layers from input to output being somatosensory neurons of the periphery and within the central nervous system (CNS), sensory processing neurons, pattern-generating premotor neurons, and motor neurons (Meng and Heckscher 2020). Neurons in different layers are connected to each other in specific patterns called circuit motifs (e.g., feed-forward, lateral inhibition, etc.), each of which performs a specific computation (Marcus et al. 2014; Braganza and Beck 2018; Luo 2021). For example, feed-forward motifs consist of one neuron type providing direct and indirect input to a second neuron type. Feed-forward circuit motifs are abundant, found in many animals (e.g., nematodes, insects, mice) and in many brain regions (e.g., somatosensory, olfactory, neocortex) where they can act as signal-onset detectors (Anton and Homberg 1999; Harris and Shepherd 2015; Schafer 2016; Wang et al. 2022). The major determinant of nervous system function is how sets of neurons are assembled into circuit motifs.
The motor circuit assembly process must be rapid, precise, and robust. Assembly starts during gastrulation, as a small pool of progenitor cells are specified. Progenitors divide tens to hundreds of times to produce a large number of many diverse neurons. Neurons are sparsely connected by synapses to form specific circuit motifs. Progenitors cannot make all neurons simultaneously, and yet, for optimal survival, the animal needs functional circuits as soon as possible. Additionally, the animal must be able to add and refine circuits over time. These challenges lead to several fundamental questions about motor circuit assembly: How many and what types of neurons does the organism make? In what order? How are neurons mapped into circuit motifs? How does an embryo robustly generate circuits, given its limited resources? What makes the process energy-efficient? What are the failure modes and recovery mechanisms?
Drosophila melanogaster as a Model to Study Motor Circuit Assembly
Until recently, studying circuit assembly had been experimentally difficult. Generally, systems with large cells (e.g., leech, lamprey) are accessible with electrodes and can be used for studies of circuit function. However, these organisms have long generation times and lack the molecular genetic tools needed to trace and manipulate stem cells and neurons. Conversely, genetic models (e.g., mice, flies) have short generation times and are amenable to genetic studies at the cell and molecular level. Electrode-based studies of circuit function in these systems, however, are challenging. Models are thus needed that permit researchers to implement circuit-level approaches while also allowing for developmental longitudinal studies. Several recent technical advances now enable the study of circuit assembly in developmental genetic model organisms. Here, we focus on one of those systems, the Drosophila larvae, which features a sizeable molecular toolkit and considerable available knowledge.
The goal of this review is to highlight current knowledge relevant to circuit assembly in Drosophila larvae, including (1) the larval body and its movements, (2) larval neurons and circuits of the motor system, and (3) how generation of neural diversity relates to circuit architecture. Separately, and as part of this collection, we also provide step-by-step protocols useful for the study of motor circuits and their assembly in Drosophila larvae (see Protocol: Imaging Neural Activity in Intact, Semirestrained Drosophila Larvae (Vasudevan et al. 2024), Protocol: Fluorescent In Situ Hybridization Chain Reaction for RNA in the Drosophila Embryonic and Larval Central Nervous System (Henderson et al. 2024), Protocol: Studying Drosophila Larval Behavior in Agarose Channels (Greaney and Heckscher 2024), and Protocol: Single-Neuron Labeling in Drosophila Using Multicolor FLP-Out (Marshall et al. 2024).
THE DROSOPHILA LARVAL BODY AND MOVEMENT
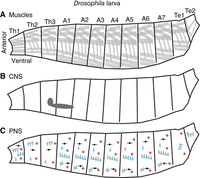
Any study of motor circuit assembly is anchored in the biology of the organism, focusing on what needs to be assembled and why. Anatomically, the Drosophila larval body is a left–right symmetrical, segmented, limbless “tube” (Fig. 1A; Hartenstein 1993). The anterior tip of the body is the first thoracic segment (i.e., Th1). It contains dark, H-shaped mouth hooks, the evolutionary remnants of the head capsule (Grimaldi and Engel 2005). In addition to segment Th1, in the anterior, there are two additional segments (Th2 and Th3). Together, Th1–Th3 compose the larval thorax, essentially its head. Adjacent to the thorax, there are seven abdominal segments (A1–A7), which can be considered the mid-body. The posterior-most region of the larval body is termed the terminus (or tail) and is composed of a few highly reduced segments (Te1–Te2). In the context of motor circuits, this segmented tube is moved during locomotion.
The organization of the Drosophila larval body. Illustrations of the Drosophila larval body are shown in the side view with anterior to the left and dorsal up. The body is left–right symmetrical and segmentally repeated. Segments are grouped into regions, and each segment within a region is numbered. Here, regions are Th (thorax), A (abdomen), and Te (terminus). (A) Organization of Drosophila muscles, with each gray box representing a muscle cell. Muscles are grouped into dorsal, lateral, and ventral sets, which repeat along the body axis. Not all muscles are shown. (B) The illustration shows the position of the larval central nervous system (CNS in gray), with brain lobes (left, circular) and nerve cord (right, elongated circle). (C) The illustration shows the larval peripheral nervous system (PNS) with different classes of primary somatosensory neurons displayed as symbols. Chordotonal neurons (vibration sensors) are shown as blue triangles. Red diamonds represent multidendritic class four neurons (nociceptors). Dorsal bipolar dendrite neurons (stretch receptors) are shown in black. The distribution of primary somatosensory neurons is identical in abdominal segments but diverges in the thorax and terminus. Not all sensory neuron types are shown.
The larval body is flexible and easily deformed. There is no hard internal or external skeleton that limits its movements. In theory, the type of movements this animal can generate is vast. However, most of the time, a larva's primary movement is peristaltic forward crawling. A more detailed look at locomotion reveals that the different regions of the larval body produce various movements. The thoracic region can perform “head sweeps” or left–right asymmetrical turns that underlie navigational decision-making (Lahiri et al. 2011; Berni 2015). The abdominal region generates peristaltic waves of movement that move the body forward or backward. The waves start from the posterior and propagate to the anterior during forward crawling, and during reverse crawling, the wave reverses direction (Heckscher et al. 2012). The terminal region initiates forward crawling in a movement sometimes called a “piston” (Heckscher et al. 2012).
Not only is the larval body important for locomotion, it also contains all of the somatosensory neurons of the peripheral nervous system (Fig. 1C). Drosophila larvae, like most animals, process a wide variety of somatosensory stimuli including self-movement, vibration, light touch, noxious temperature, and mechanical stimuli (Tracey et al. 2003; Yan et al. 2013; Wreden et al. 2017). Somatosensory stimuli are used to refine movement and for action selection (e.g., initiating an escape roll, triggering, and avoidance hunch) (Hwang et al. 2007; Jovanic et al. 2016).
DROSOPHILA LARVAL NEURONS AND CIRCUITS
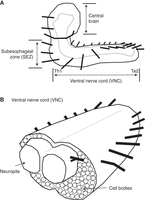
Also important for the study of motor circuit assembly is an understanding of the “parts list”; i.e., the components that comprise circuits. Like the larval body, the Drosophila larval CNS (Fig. 1B) is left–right symmetrical and segmented (Fig. 2A,B). Generally, there is good correspondence between the body and CNS regions (e.g., the neurons found in the A1 segment of the CNS monitor and control the A1 segment of the body). Each side (left or right) of a body segment is termed a “hemisegment.” Each side of the CNS segment is called a “hemineuromere.” The A1 body segment has been considered representative of other abdominal segments, but notable differences exist between the thorax and terminus (Rickert et al. 2011; Heckscher et al. 2014). In an A1 hemisegment, 30 muscles are grouped into three major sets: dorsal, lateral, and ventral (Fig. 1A; Bate 1990). The recruitment of muscles during some behaviors has been characterized (Heckscher et al. 2012; Zarin et al. 2019; Liu et al. 2023). The 35 motor neurons of each hemisegment have been mapped to their corresponding muscle at single-cell resolution (Landgraf et al. 1997). Per A1 hemineuromere, in addition to the 35 motor neurons are approximately 270 distinct interneurons. Here, interneuron refers to any neuron that is not a motor neuron. Each left–right pair of interneurons in the A1 neuromere has unique morphology (Bossing and Technau 1994; Bossing et al. 1996; Schmidt et al. 1997; Schmid et al. 1999; Rickert et al. 2011). Additionally, there are 43 different sensory neurons in each abdominal hemisegment (Fig. 1C; Bodmer and Jan 1987; Grueber et al. 2003), some of whose recruitment patterns and role in behavior have been characterized (e.g., He et al. 2019; Vaadia et al. 2019).
The anatomy of the Drosophila larval central nervous system (CNS). (A) An illustration of the Drosophila larval CNS is shown in a side view, anterior to the left and dorsal to the top. The dashed line represents the border of the neuropile. Each black line projecting from the CNS represents a nerve root. There is one nerve root per segment. The three major CNS regions are labeled (central brain, SEZ [subesophageal zone], VNC [ventral nerve cord]). Motor circuits are housed in the ventral nerve cord. (B) A diagram of a cross-section of the VNC is shown, anterior to the left. The neuropile is an axon-, dendrite-, and synapse-rich region. Cell bodies are peripheral to the neuropile. Each black line projecting from the CNS represents a nerve root.
A Drosophila larval connectome (Ohyama et al. 2015) provides detailed anatomical access to larval circuitry. The nerve cord comprises more than 10,000 neurons joined by millions of synapses, each of which can be visualized in this data set. Although the connectome is an image of only one larval time point, as the larvae grow, it is believed that additional neurons are not added to the larval network. Instead, neurons and muscles grow and add synapses between already established partners (Gerhard et al. 2017). Thus, this connectome is an excellent resource for circuit description. Many circuits have been described using connectomics and other tools, and we refer the reader to comprehensive recent reviews on the topic (Kohsaka et al. 2012; Clark et al. 2018; Eschbach et al. 2020). A major question, however, is to what extent neural cell fate specification also dictates morphology and connectivity and therefore circuit motif topology.
DROSOPHILA NEURAL STEM CELLS AND THE GENERATION OF NEURAL DIVERSITY
Fundamentally, the study of circuit development aims to determine how networks form. One useful way to parse the problem is to consider that circuit networks are composed of nodes (neuronal somata) and edges (connections between nodes formed by axons, dendrites, and synapses). Generally, in developmental neurobiology, there is a solid understanding of the origins of neural diversity, which is important because it tells us about how many nodes and of what type are available to the network.
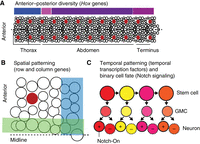
Briefly, four major processes account for generating the diversity of neural somata in the Drosophila nerve cord (Fig. 3). In Drosophila, neuronal stem cells are called “neuroblasts.” Nerve cord neuroblasts are among the most intensely studied and best-characterized neuronal stem cells (Goodman and Doe 1993; Broadus et al. 1995; Birkholz et al. 2013, 2015) in part due to their stereotyped morphology. All neurons that will generate larval CNS circuits are born during embryogenesis from nerve cord neuroblasts. Neuroblasts in different regions along the anterior–posterior axis are genetically nonidentical, partially due to the activity of Hox transcription factors (Fig. 3A; Estacio-Gómez and Díaz-Benjumea 2014). In each segment, there are left–right pairs of neuroblasts that comprise 30 different classes (Fig. 3B). One set of 30 neuroblasts generates approximately 300 neurons (Bossing et al. 1996; Schmidt et al. 1997; Heckscher et al. 2014). In embryos, each neuroblast is named for its position within a row–column array (e.g., NB3-3 is in the third row, third column) (shown in red in Fig. 3A,B; Doe 1992; Broadus et al. 1995). We describe NB3-3 lineage in more detail below. When these same neuroblasts are found at larval stages, they are given different names (see Lacin and Truman 2016 for details). In embryos, each neuroblast attains a unique identity based on spatial patterning. Spatial patterning relies on the combinatorial code of “row” and “column” transcription factors (Fig. 3B; Skeath 1999; Skeath and Thor 2003). The set of progeny produced by a single stem cell is unique and stereotyped and is termed a lineage. Neural lineages comprise groups of nonidentical neurons (Fig. 4A; Hartenstein and Stollewerk 2015). Over time, each neuroblast generates different neurons (e.g., first-born, second-born) in a process called temporal patterning. Temporal patterning is implemented by the activity of temporal transcription factors (Fig. 3C; Isshiki et al. 2001; Pearson and Doe 2003; Grosskortenhaus et al. 2006; Tran and Doe 2008). An additional layer of diversity is achieved at the level of neuroblast daughter cells. Typically, in a “type 1” division, a neuroblast generates a ganglion mother cell (GCM), which divides to give rise to a Notch-On daughter and a Notch-Off daughter (Fig. 3C). Daughters take on different binary cell fates (Skeath and Doe 1998; Truman et al. 2010). Thus, one neuroblast can give rise to two hemilineages—one comprised of Notch-On daughters and the other comprised of Notch-Off daughters. Neurotransmitter phenotypes are well correlated with hemilineages (Lacin and Truman 2016). Thus, in the Drosophila motor system, we currently have a good understanding of how nodes are formed. In contrast, we know much less about how edges are formed.
The generation of neural diversity in Drosophila nerve cord. (A) Neuroblasts are arrayed along the anterior–posterior axis. Each circle represents one neuroblast. The midline is represented by a dashed line. A representative neuroblast, NB3-3, is shown in red to highlight the left–right symmetrical and serially homologous organization of the nerve cord. An example of a nonrepeating unit, a hemineuromere, is shown in the box at the bottom left and is enlarged in B. Above the nerve cord, colored bars show approximate zones of Hox gene expression. Regions of the nerve cord are labeled below. (B) A single hemineuromere (box in A) is shown. Each neuroblast is distinct due to row and column gene expressions, which are represented by blue and green stripes. (C) The illustration shows neuroblast lineage progression. Each circle represents one cell, and each arrow represents a cell division. The top row contains neuroblasts that divide to self-renew and generate a ganglion motor cell (center row). (Bottom row) Ganglion mother cells (GMCs) divide asymmetrically to generate two neurons. Neurons become different from each other based on Notch signaling, with cells labeled with “+” representing Notch-On neurons and the others representing Notch-Off neurons. Temporal transcription factor expression in neuroblasts is dynamic, changing on each division, with each color representing a different temporal transcription factor. However, temporal transcription factors are maintained in the progeny generated at specific birth times (they keep their color).
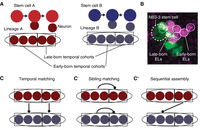
Stem cell lineages are organized into temporal cohorts in Drosophila. (A) In this illustration, stem cell A (red) and stem cell B (blue) generate two distinct lineages (ovals) of neurons. Both lineages are subdivided into temporal cohorts (boxes). Each circle represents a cell, with arrows representing cell division. (B) An image shows early-born and late-born Even-skipped (Eve)-expressing lateral (EL) neurons (small circles, each ∼3 µm in diameter) from the NB3-3 stem cell (large dashed circle at left). This late-stage embryo was stained with an anti-Even-skipped antibody (magenta) to label all EL neurons (small circles) and costained with anti-GFP (green). In this embryo, GFP expression is being driven by an enhancer, “11F02” (genotype: 11F02-GAL4 > UAS-nls-GFP). (C) Lineages can be connected to each other in different patterns. Symbols are as in A and, additionally, the line with a dot on the end represents synapses between neurons of one temporal cohort and another temporal cohort. In temporal matching, early-born neurons and late-born neurons from different lineages synapse with each other. (C′) In sibling matching, neurons from the same lineage synapse with each other. (C″) In sequential assembly, neurons from a late-born temporal cohort in one lineage synapse with neurons from an early-born temporal cohort in a second lineage. Because neurons make multiple connections, these models are not mutually exclusive.
In the past decade, there have been major advances in our understanding of circuit formation. Fundamentally, three stem cell-to-circuit patterns have been described in the Drosophila system and others. The first pattern is “temporal matching” (Fig. 4C), where early-born neurons from one lineage wire together with early-born neurons from a second lineage (McLean et al. 2007; Fetcho and McLean 2010; Pujol-Martí et al. 2012; Bagnall and McLean 2014; Pujala and Koyama 2019; Mark et al. 2021; Goldblatt et al. 2023). The second pattern is “sibling matching” (Fig. 4C′), where sister neurons from one lineage wire together (Yu et al. 2009; Wang et al. 2022). The third pattern is “sequential assembly” (Fig. 4C″), where output neurons of a circuit are born before their input neurons (Wang et al. 2022). In these cases, one shared aspect of wiring is that circuits are assembled from small sets (approximately five to six) of neurons born from a single stem cell within a tight time window (Wang et al. 2022). These sets of neurons are developmental units called “temporal cohorts” (Wreden et al. 2017).
The discovery of temporal cohorts was seeded by an observation from an enhancer expression screen (Manning et al. 2012). “11F02-GAL4” is a 3-kb DNA fragment that drives the expression of GAL4. Using 11F02-GAL4 to drive a UAS-GFP reporter, we found GFP expression in half of the neural progeny made by the NB3-3 stem cell. In segment A1, NB3-3 stem cell lineage produces 11 Even-skipped (Eve)-expressing lateral (EL) neurons (Fig. 4B, magenta; Schmidt et al. 1997; Schmid et al. 1999). Curiously, halfway through development “11F02” is expressed in the NB3-3 stem cell and all late-born EL neurons (and a handful of other neurons) (Fig. 4B, green). Thus, 11F02 expression subdivides neurons of the NB3-3 lineage into early-born and late-born sets. Our laboratory used the 11F02 enhancer fragment to build genetic tools to probe the function of early-born and late-born EL neurons separately. Early-born EL neurons contribute to a circuit that processes mechanical stimuli and triggers escape rolling. In contrast, late-born EL neurons contribute to a circuit that processes proprioceptive stimuli and regulates body posture (Wreden et al. 2017). We also reconstructed all synaptic inputs onto early-born and late-born EL neurons in a single segment (A1) of the nerve cord using connectomics. There were statistically significant similarities in connectivity for early-born EL neurons and separately for late-born EL neurons, and statistically significant differences between early-born and late-born EL neurons (Wang et al. 2022). Thus, circuit membership of neurons in the NB3-3 lineage is similar over time, until it changes dramatically within one cell division. These changes in wiring correspond to the developmental unit called a temporal cohort.
Since the initial description of temporal cohorts in the NB3-3 lineage, other examples have been discovered in the A1 segment of the Drosophila nerve cord (Meng et al. 2019, 2020; Mark et al. 2021; Wang et al. 2022). Temporal cohorts are important developmental units relevant to circuit wiring (Meng and Heckscher 2020). Furthermore, temporal cohorts are found in the adult Drosophila nerve cord (Baek and Mann 2009), and likely in other parts of the Drosophila CNS (e.g., mushroom body, central complex) and vertebrate spinal cord (Stam et al. 2012). This demonstrates that temporal cohorts are abundant. One consistent theme is that circuit motifs are assembled from a limited number of temporal cohorts from different stem cell lineages (Meng and Heckscher 2020).
CONCLUDING REMARKS
Motor circuits are an ancient part of the nervous system that perform fundamentally similar computations in all animals. Research has uncovered striking parallels in motor circuit development between the vertebrate spinal cord and the Drosophila nerve cord (Wreden et al. 2017; Jay and McLean 2019; Catela and Kratsios 2021). These observations suggest that the development of motor circuit assembly could be conserved across phyla, and they motivate continued research in Drosophila.
Although progress in understanding motor circuit assembly in Drosophila larvae has been made recently, many questions remain. We understand only a fraction of the relationships between stem cell lineage and circuits. Outstanding questions include the following: What other stem cell relationships will be found? How do stem cell–circuit relationships vary, along the anterior–posterior axis, with life stage, and among closely related organisms? On the molecular and cellular levels, how are relationships constructed? What are the roles of stem cell cues (e.g., inherited chromatin state, transcription factor expression), neural cues (e.g., cell surface molecules), and environmental cues (e.g., signaling gradients, physical interactions)? In terms of perturbative analyses, what perturbations is the system robust to? When the system is disrupted, where and how does it break? Are there synergistic interactions among perturbations? Are there specific repair mechanisms, or are normal developmental programs reused? These and related questions will keep investigators in the field occupied for the coming decade.
Footnotes
-
From the Drosophila Neurobiology collection, edited by Bing Zhang, Ellie Heckscher, Alex C. Keene, and Scott Waddell