Measuring Apoptosis: Caspase Inhibitors and Activity Assays
- 1Department of Life Sciences, New York Institute of Technology, Old Westbury, New York 11568;
- 2Department of Immunology, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105
Abstract
Caspases are proteases that initiate and execute apoptotic cell death. These caspase-dependent events are caused by cleavage of specific substrates that propagate the proapoptotic signal. A number of techniques have been developed to follow caspase activity in vitro and from apoptotic cellular extracts. Many of these techniques use molecules that are based on optimal peptide motifs for each caspase and on our understanding of caspase cleavage events that occur during apoptosis. Although these approaches are useful, there are several drawbacks associated with them. The optimal peptide motifs are not unique recognition sites for each caspase, so techniques that use them may yield information about more than one caspase. Furthermore, caspase cleavage does not take into account the different caspase activation mechanisms. Recently, probes having greater specificity for individual caspases have been developed and are being used successfully. This introduction provides background on the various caspases and introduces a set of complementary techniques to examine the activity, substrate specificity, and activation status of caspases from in vitro or cell culture experiments.
INTRODUCTION TO CASPASES
Every cell and every organism that undergoes apoptotic cell death uses caspases. This family of proteases plays critical roles in inflammation and cell death, and is essential for the initiation and execution of apoptosis. Cellular signals, such as death ligands or mitochondrial outer membrane permeabilization (MOMP), activate the first of the two classes of caspases: Initiator caspases. Activated initiator caspases either cleave and activate executioner caspases, or cleave other proteins that transmit the apoptotic signal to the executioner caspases. Executioner caspases are responsible for cleaving a myriad of substrates, some of which mediate the apoptotic phenotype in dying cells.
Caspases are cysteine-dependent aspartic-acid-directed proteases. Caspase substrates are cleaved at the carboxy-terminal peptide bond of the aspartic acid residue in the peptide motif, causing a loss or gain of function of the target protein, generally favoring the proapoptotic signaling pathway. The first mammalian caspase to be identified was caspase-1. It was detected by its ability to process the inflammatory cytokine interleukin 1β (Cerretti et al. 1992; Thornberry et al. 1992). The cleavage site in interleukin 1β after aspartic acid residue 116 was the first hint regarding the specificity of caspase cleavage motifs. Since this initial discovery, the caspase family has grown substantially (Kumar 2007).
ACTIVATION OF CASPASES
Caspases become activated in one of the two ways, depending on whether they are initiator or executioner caspases. Initiator caspases (e.g., caspase-2, -8, -9, and -10) are activated by dimerization on large multiprotein complexes. Caspase-8 and -10 activation occurs via the death-inducing signaling complex (DISC) that is formed in response to death ligands (Medema et al. 1997; Sprick et al. 2002). Caspase-9 activation occurs on the apoptosome, which is formed when cytochrome c and adenosine triphosphate bind to apoptotic protease activating factor-1 (APAF-1) causing oligomerization (Li et al. 1997). Caspase-2 is activated by the PIDDosome, a poorly understood complex containing the adaptor proteins p53 inducible protein with a death domain (PIDD) and RIP-associated ICH/CED-3 homologous protein with a death domain (RAIDD) (Tinel and Tschopp 2004). In each case, the induced dimerization event results in the formation of an active site on one or both of the caspase monomers. Once dimerized, the caspase is able to cleave target substrates (Boatright et al. 2003).
Executioner caspases (e.g., caspase-3, -6, -7) exist as inactive dimers that are activated upon proteolytic cleavage of both monomers at loops containing specific cleavage sites (Berger et al. 2006a; Denault et al. 2006; Edgington et al. 2012). Cleavage of these loops results in structural changes that serve to form the active sites, allowing the executioner caspases to cleave downstream substrates (Chai et al. 2001; Riedl et al. 2001). Activated executioner caspases can be detected using antibodies that recognize caspase proforms or cleavage fragments (see Protocol: Detection of Caspase Activity Using Antibody-Based Techniques [McStay and Green 2014a]).
During apoptosis, all caspases undergo cleavage at similar sites located between the large and small catalytic domains. In addition, in the case of initiator caspases, there is often additional cleavage between the pro-domain and the catalytic domains. These cleavage events are mediated by auto-cleavage and other caspases. The function of these cleavage events differs slightly between initiator and executioner caspases. Cleavage of initiator caspases-2 and -8 stabilizes the enzyme (Baliga et al. 2004; Oberst et al. 2010). In the case of caspase-9, cleavage dissociates the enzyme from the activation platform, whereby the caspase is replaced by another pro-caspase-9 molecule in a “tick-over” mechanism (Malladi et al. 2009). With executioner caspases, cleavage is necessary and sufficient to induce their activation (Riedl and Shi 2004). This has been shown by comparing inactive and active caspase-7 structures, which revealed that cleavage of its loops freed up the caspase active site (Chai et al. 2001; Riedl et al. 2001).
CASPASE SUBSTRATES
Using combinatorial peptide libraries and purified recombinant caspases, characterization of caspase substrates has revealed the presence of a four-amino-acid consensus motif upstream of the cleavage site that is recognized for substrate proteolysis. The motif is designated as P1, P2, P3, and P4, where P1 is the amino acid closest to the cleavage site. Positions P2 and P4 contain amino acids with similar properties and positions P1 and P3 are held by specific amino acids. Based on structural studies of caspase active sites, caspases-3, -6, and -7 only accommodate small residues such as alanine or valine at P2. Initiator caspases are able to tolerate bulkier residues at this position. The P3 site is occupied by a conserved arginine residue that favors glutamate binding in the substrate pocket. Caspases-2, -3, and -7 accommodate an aspartate residue at P4 with high specificity, while in the other apoptotic caspases the P4 site tolerates a variety of amino acids. The P4 site of caspase-1 can accommodate large hydrophobic amino acids, such as tryptophan (Chereau et al. 2003). The vast majority of caspase substrates contain an aspartate residue at the P1 position. Cleavage occurs by hydrolysis of the peptide bond following this residue. Table 1 contains a summary of the motif preferences of each caspase based on structural features. These are merely preferences and do not imply that other sites cannot be cleaved by these enzymes. Indeed, misconceptions surrounding these preferences have led to misinterpretations in the literature, as discussed in more detail below.
Caspase cleavage motifs
Approximately 1000 caspase substrates have been identified (Mahrus et al. 2008; Crawford et al. 2012; Shimbo et al. 2012) with functions that encompass many cellular processes. Poly-ADP-ribose polymerase was the first apoptotic caspase substrate to be identified (Tewari et al. 1995). Recently, caspase substrates have been identified by large-scale proteomic or in silico approaches (Table 2). Caspase cleavage of a substrate can yield one of the several outcomes: The substrate loses function, gains a function, changes its location, or is subjected to altered regulation. These changes in the substrate are subsequently responsible for the apoptotic phenotype—for example, phosphatidylserine exposure on the external face of the plasma membrane, loss of mitochondrial membrane potential, and chromatin condensation. Proteomic approaches have been used to determine large numbers of caspase substrates during apoptosis (Van Damme et al. 2005; Mahrus et al. 2008). The substrates identified by these methods have been classified by annotation into specific pathways or functions, giving us some idea of processes altered by active caspase cleavage. These putative substrates must be validated using some kind of experimental screen, such as is described in Protocol: Verification of a Putative Caspase Substrate [McStay and Green 2014b]). On a smaller scale, putative caspase cleavage motifs in proteins can be identified by in silico prediction models (Scott et al. 2008).
Caspase substrate databases and substrate prediction servers
The consensus substrate motifs have been used to generate short peptide-based inhibitors and substrates for each known caspase (Talanian et al. 1997; Thornberry et al. 1997; Garcia-Calvo et al. 1998). These reagents, which are commercially available, have been used extensively to test the involvement of caspases in apoptosis. Despite their utility, these reagents lack the selectivity they are claimed to have for individual caspases in whole cell and in vivo assays where multiple caspases are present (Garcia-Calvo et al. 1998; Ekert et al. 1999), as well as in some in vitro assays (Berger et al. 2006b; McStay et al. 2008; Pereira and Song 2008; Benkova et al. 2009). This lack of specificity may be attributed to the expression, activation, substrate specificity, and/or activity of caspases that prevent a substrate-based tool from selecting for a single caspase when multiple caspases are present. Let us look at these reasons more closely.
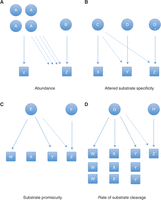
Caspase protein expression levels shows cell-type variability, as was showed in a study that measured caspase protein levels in a selection of cancer cell lines (Svingen et al. 2004). Because there are no comprehensive reports on protein expression of caspases in tissues or cell lines, one should empirically determine caspase protein expression relative to a standard amount of protein or a housekeeping protein in the cell line or tissue of interest. Problems can arise when using a peptide-based substrate to study a caspase with specific-substrate cleavage ability, if that caspase is present at lower levels than another caspase that is both more highly expressed has promiscuous substrate cleavage ability. The result may be that the more abundant caspase is responsible for a significant amount of substrate cleavage although the substrate was designed to probe the activity of a different caspase (Fig. 1A).
Properties of caspases that confound interpretation of assays using substrate motif-based activity probes and inhibitors. (A) Caspase A has a greater expression level than caspase B. (B) Caspase D has altered substrate specificity when in an intermediate activation step (D*) compared to the fully mature enzyme. (C) Caspase E is able to cleave many different substrates while caspase F cleaves only one particular substrate. (D) Caspase G is able to cleave many substrates in the same amount of time caspase H cleaves one substrate.
The substrate specificity of some caspases can change depending on the activation state. Since there are two caspase activation mechanisms, inference of activity by use of substrate-based tools also needs to consider the mechanism and state of activation. As was discussed previously, initiator caspases go through a multistep process in which one or both active sites can be in the active conformation. In the case of caspase-8, these different forms display different substrate preferences (Hughes et al. 2009). With executioner caspases, cleavage of a specific loop on both monomers is required to induce a conformational change in the enzyme that makes the active site available to the whole repertoire of substrates at the same time (Fig. 1B).
Initiator caspases are activated in large complexes in response to a proapoptotic stimulus. These caspases do not have to cleave their limited suite of substrates at a rapid rate. Executioner caspases, on the other hand, cleave on the order of a thousand substrates and must do so quickly to ensure the cell dismantles itself in an orderly and timely fashion and is engulfed by phagocytic cells to avoid activation of inflammation. Therefore, the degree of substrate specificity—limited for initiator caspases and broad for executioner caspases—can affect experiments using peptide-based substrate probes. The promiscuity of an executioner caspase might swamp the signal of an initiator caspase (Fig. 1C).
As noted, caspases cleave substrates at different rates. For example, caspase-3 is much more active than any other caspase at cleaving its favored substrate (Stennicke et al. 2000). This variation in cleavage rate can also confound interpretation of assays using putative caspase-specific substrates when multiple caspases are present (Fig. 1D).
INHIBITORS OF CASPASES
Caspase inhibitors are available as either pan-caspase or caspase-specific inhibitors. Depending on the assay used to measure apoptosis, these inhibitors will not always prevent cell death. This may be due to toxicity associated with caspase inhibitors, cell death that is caspase independent, or ineffective employment of the caspase inhibitor. The most common caspase inhibitors are short peptides with a moiety that covalently binds to the catalytic cysteine. These moieties include fluoromethyl ketone (FMK), chloromethyl ketone and 2,6-difluorophenoxymethyl ketone (OPH). FMK moieties are metabolized to fluoroacetate, an inhibitor of aconitase, which leads to mitochondrial toxicity in vivo and possibly necrotic features in vitro (Van Noorden 2001; Chauvier et al. 2007). The OPH moiety is less toxic and tolerated by many different cell types and is less toxic in vivo (Chauvier et al. 2007).
Caspase inhibitor studies have revealed two types of death pathways. First, initiation of the intrinsic pathway of apoptosis in the presence of caspase inhibitors does not result in apoptotic features, but cells lack clonogenic survival. This phenomenon occurs because mitochondrial outer membrane permeabilization (MOMP) is a point from which cells often cannot recover, cytochrome c is released into the cytosol, and mitochondrial-dependent energy production is severely impaired. Under most circumstances, cells are unable to circumvent this problem and will eventually die due to depletion of ATP and decreased mitochondrial functions necessary for survival, even if caspases downstream from MOMP are inhibited. Second, cells exposed to tumor necrosis factor-α in the presence of caspase inhibitors do not undergo apoptosis; instead they engage a pathway called necroptosis. Inhibition of the catalytic activity of caspase-8 prevents cleavage of receptor interacting protein-1 (RIPK1) kinase, which engages RIPK3 kinase to initiate downstream signaling that results in caspase-independent cell death (Oberst et al. 2011). Finally, to inhibit apoptosis, caspase inhibitors need to gain access to the cytosol and be present at an appropriate concentration. By monitoring characteristic executioner caspase-dependent events, such as caspase-3 activity assays or cleavage, it can be seen whether an appropriate concentration of inhibitor was used. To claim that a cell death pathway acts independently of caspases, these caspase-dependent events must be completely inhibited as the cell proceeds to die. Therefore, even if cells are treated with caspase inhibitors, it does not mean that they will necessarily survive over the long term.
NONCASPASE PROTEASES INVOLVED IN CELL DEATH
Proteases other than caspases have been implicated in some cell death paradigms that can impinge on the canonical apoptotic pathway. These include the serine protease family, of which one directly engages apoptosis. Granzyme B is a serine protease secreted from cytotoxic cells to kill infected or malignant cells. Granzyme B enters the target cell where it cleaves and activates caspase-3 directly, leading to apoptosis (Darmon et al. 1995). Also, human Granzyme B cleavage of BID causes MOMP and apoptosis (Cullen et al. 2007). Cathepsins are another family of proteases postulated to be involved in apoptosis. These are cysteine proteases that reside mainly in the lysosome and are mostly responsible for proteolysis in lysosomal degradation pathways. In some forms of cell death, cathepsins are released from lysosomes into the cytosol, where they can cleave a variety of substrates (Repnik et al. 2012). There are many cathepsin-specific inhibitors, but these proteases are also sensitive to caspase inhibitors like zVAD-FMK (Rozman-Pungercar et al. 2003). Calpains are calcium-activated cysteine proteases that have been implicated in some apoptotic pathways (Storr et al. 2011). Some inhibitors of cathepsins also inhibit calpains, including zVAD-FMK (Rozman-Pungercar et al. 2003). Unlike calpains and cathepsins, Granzyme B is not inhibited by any known caspase inhibitors.
ASSAYS TO STUDY CASPASES
Caspases degrade specific substrates after apoptosis or inflammation is initiated. Therefore, tracking caspase activity during these cellular processes should reveal the roles caspases are playing in specific signal transduction pathways. The conventional way to follow caspase activity is by monitoring cleavage of either model peptide substrates or physiological substrates of the enzyme. These assays can be performed on apoptotic cell extracts, in vitro-activated caspases (see Protocol: Preparation of Cytosolic Extract and Activation of Caspases by Cytochrome c [McStay and Green 2014c]), or on purified caspases. Using the first two systems, the contributions of multiple caspases found within a cell at a given time can be analyzed. Working with a purified caspase permits more detailed analysis of the function of that caspase. The caspases involved in specific apoptosis scenarios can be identified using substrate-based inhibitor studies (see Protocol: Assaying Caspase Activity In Vitro and Protocol: Identification of Active Caspases Using Affinity-Based Probes [McStay and Green 2014d,e]). Although the available caspase inhibitors show limited selectivity for an individual caspase, by performing complementary assays the identification of an activated caspase in the apoptotic cascade can be confirmed. These assays also aid in determining the mechanisms of caspase activation that differ among the classes of caspases and among individual caspases from the same class. The combination of these two properties of caspases is important for further understanding the contributions caspases make in many different apoptosis scenarios.
When caspase cleavage motif-based tools are used, complementary assays must be performed to validate the involvement of a suspected caspase. Caspase-3 cleaves multiple caspase cleavage motifs, and analysis of the contribution of this caspase should be determined by using the preferred caspase cleavage motif DEVD. Experiments using inhibitors must be supported with genetic approaches that lower the amount of caspase, either by using interfering RNA or cells derived from knockout animals that lack the specific caspase. Such complementary approaches have been used to identify the emerging role of caspase-8 as an activator of interleukin 1β following a variety of extracellular inflammatory stimuli (Maelfait et al. 2008; Bossaller et al. 2012; Gringhuis et al. 2012; Vince et al. 2012). These substrate motif-based reagents still represent the only source of commercially available substrates or inhibitors. A new series of activity-based probes based on natural and nonnatural amino acids have been generated, which show enhanced specificity for individual caspases in some cases (Berger et al. 2006a; Edgington et al. 2009, 2012). The activity-based probe developed for caspase-8 cross-reacts with caspase-3. The probe developed for caspase-9 cross-reacts with caspase-3 and -8. Further refinement of these probes and ultimately the design of true caspase-specific probes will require an approach that integrates information about structure and mechanisms of activation.
USE OF COMMERCIAL KITS TO MONITOR CASPASES
Many life science companies sell kits that provide all the necessary reagents to monitor caspases in models of apoptosis. Generally, these kits supply buffers for cell lysis and caspase assays, along with a fluorogenic substrate and inhibitors to the caspase in question. Some kits are marketed for individual caspases and others are for multiple caspases. The kits putatively for individual caspases generally contain reagents for that particular caspase, and thus have more limited utility. The kits that have broad specificity contain substrate-based reagents of all caspases and therefore allow for direct comparison among substrates, inhibitors, and caspases at the same time. It is also possible to purchase recombinant caspases either alone or as part of some assay kits. These may be worthwhile if only a limited numbers of assays are planned. For extensive studies with recombinant caspases, it is more economical to prepare recombinant caspases in-house. This rule may also hold true for caspase inhibitors and fluorogenic substrates, which may be synthesized at a core facility when a large number of experiments are planned.
CONCLUSION
The biochemical steps of caspase activation and cleavage of caspase substrates have been extensively studied since the first apoptotic caspase and substrate were identified. These studies have generated a variety of reagents and assays to interrogate the roles of caspases and their substrates in many paradigms of apoptosis. These reagents include small molecule substrates and inhibitors that can be used to determine activation of caspases and cleavage of substrates after proapoptotic stimuli. In isolation, these assays provide clues to the involvement of a particular caspase in a particular apoptosis scenario. However, these assays by themselves do not provide all the necessary information about the activation of a caspase or whether a single caspase was responsible for the observed apoptotic process. By combining these reagents and assays, while complementing them with more recently developed experimental tools, a more complete understanding of which caspases are involved and how the caspases are activated in apoptotic pathways can be achieved.
- © 2014 Cold Spring Harbor Laboratory Press