Generation and Selection of Phage Display Antibody Libraries in Fab Format
- Department of Immunology and Microbiology, The Herbert Wertheim UF Scripps Institute for Biomedical Innovation & Technology, University of Florida, Jupiter, Florida 33458, USA
- ↵1Correspondence: rader33458{at}gmail.com
Abstract
Monoclonal antibodies (mAbs) have exceptional utility as research reagents and pharmaceuticals. As a complement to both traditional and contemporary single-B-cell cloning technologies, the mining of antibody libraries via display technologies—which mimic and simplify B cells by physically linking phenotype (protein) to genotype (protein-encoding DNA or RNA)—has become an important method for mAb discovery. Among these display technologies, phage display has been particularly successful for the generation of mAbs that bind to a wide variety of antigens with exceptional specificities and affinities. Rather than multivalent whole antibodies, phage display typically uses monovalent antibody fragments, such as “fragment antigen binding” (Fab), as the format of choice. The ∼50-kDa Fab format consists of four immunoglobulin (Ig) domains on two polypeptide chains (light chain and shortened heavy chain), and exhibits its antigen binding site in a natural configuration found in bivalent IgG and other multivalent Ig molecules. The Fab fragment has a high melting temperature and a low tendency to aggregate, and can be readily converted to natural and nonnatural Ig formats without affecting antigen binding properties, which has made it a favored format for phage display for more than three decades. Here, I briefly summarize some of the approaches used for the generation and selection of phage display antibody libraries in Fab format, from human and nonhuman antibody repertoires.
MONOCLONAL ANTIBODIES
Beyond their use as research reagents, monoclonal antibodies (mAbs) have found wide applications as diagnostic, preventative, and therapeutic biologics in human and veterinary medicine (Mullard 2021). The success of the antibody molecule as a pharmaceutical is, in one respect, grounded in its natural properties—specifically, its high affinity, specificity, and stability—along with its long circulatory half-life and its ability to bridge the adaptive and innate immune system through linking recognition and response. In another respect, its natural modularity has rendered it highly compatible with manipulation by protein engineering (Chiu et al. 2019).
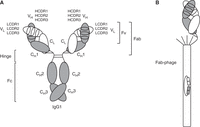
The most common natural format of mAbs is the 150-kDa immunoglobulin G1 (IgG1) molecule (Fig. 1A), which consists of two identical 25-kDa light (L) chains and two identical 50-kDa heavy (H) chains (Saphire et al. 2001; Vidarsson et al. 2014). Each chain (light and heavy) contains a variable (V) domain (VL and VH, respectively), followed by one constant domain in the light chain (CL) and three constant domains in the heavy chain (CH1, CH2, and CH3). These four polypeptide chains assemble in a Y-shaped configuration that is stabilized by four disulfide bridges in the hinge region. The two arms of the Y are known as “fragment antigen binding” (Fab) fragments, and consist of a heterodimer of the light chain (VL-CL) and the N-terminal half (VH-CH1) of the heavy chain. The stem of the Y is known as the “fragment crystallizable” (Fc) fragment, and consists of a homodimer of the C-terminal half (CH2-CH3) of the heavy chains. The N-terminal portion of the Fab, the VL and VH heterodimer that is also known as the “fragment variable” (Fv) fragment, contains the antigen binding site, or paratope. The Fv is encoded by the VL and JL genes in the κ or λ light chain locus, and by the VH, D, and JH genes in the heavy chain locus. In the human genome, the κ and λ light chain loci on chromosomes 2 and 22, respectively, comprise 44 functional Vκ, five functional Jκ, 37 functional Vλ, and seven functional Jλ genes (Watson et al. 2015). The heavy chain locus on chromosome 14, on the other hand, comprises 44 functional VH, 25 functional D, and six functional JH genes (Matsuda et al. 1998). During the initial stages of B-cell maturation in the bone marrow, VL-JL and VH-D-JH genes somatically rearrange to create functional light and heavy chain genes. These have hypervariable amino acid sequences, known as light and heavy chain complementarity determining region 3 (LCDR3 and HCDR3), encoded at the deliberately imprecise VL-JL and VH-D-JH fusion sites, respectively. Other regions with highly variable amino acid sequences are encoded by the VL (LCDR1 and LCDR2) and VH (HCDR1 and HCDR2) genes, and additional diversification in the variable domains is achieved by somatic hypermutation at later stages of B-cell maturation in the blood (Rajewsky 1996). The variable Ig domain comprises two antiparallel β sheets that interact through a hydrophobic core and are connected by a disulfide bridge. This configuration provides a highly stable scaffold that displays the CDRs as protruding β loops. In doing so, the variable Ig domain combines high amino acid sequence variability with high stability. The six CDRs of the Fv fragment essentially define the paratope that binds an epitope on the antigen with typically high specificity and affinity. In addition, the presence of two Fab arms on each IgG affords an avidity gain through bivalent binding.
IgG1, Fab, and Fab-phage. (A) Depiction of an ∼150-kDa IgG1 molecule with the two ∼25-kDa light chains shown in white and the two ∼50-kDa heavy chains shown in gray. Each of these chains (light and heavy) comprises an N-terminal variable domain (VL or VH, respectively), each having three complementarity determining regions (CDRs), followed by one constant domain in the light chain (CL) and three constant domains in the heavy chain (CH1, CH2, and CH3). VL/VH and CL/CH1 heterodimerize, and CH3/CH3 homodimerizes. The glycosylated CH2 domain does not dimerize. The hinge region between CH1 and CH2 includes four interchain disulfide bridges: two between the heavy chains and two between heavy and light chains. The VL/VH heterodimer is also known as “fragment variable” (Fv), and the combined VL/VH and CL/CH1 heterodimer is known as the “fragment antigen binding” (Fab). The hinge region connects the two Fab arms of the IgG1 molecule to the “fragment crystallizable” (Fc). (B) Depiction of a Fab-displaying filamentous phage in phagemid-based phage display systems. Note the preserved C-terminal disulfide bridge between the light chain and heavy chain fragments. The Fab is fused to the N terminus of a C-terminal fragment of minor coat protein pIII and forms, together with four wild-type pIII copies, the spiky cap of the filamentous phage particle. The phagemid encapsulated by the filamentous phage particle encodes the displayed Fab, affording the physical linkage of genotype and phenotype.
The antibody molecule is a unique protein that combines high variability in the primary structure with high stability of the secondary, tertiary, and quaternary structures. Taking advantage of the natural modular stability of the antibody molecule, antibody fragments can be used to engineer and evolve its affinity and specificity. The 50-kDa Fab fragment is particularly robust, with high solubility and dispersity, and a melting temperature (Tm) of ∼75°C (Garidel et al. 2020). It can be expressed in Escherichia coli, which is a requisite for phage display, is readily convertible to IgG1 and other natural and nonnatural Ig formats without affecting antigen binding properties, and is the modality of several therapeutic mAbs, including abciximab (Reopro) and ranibizumab (Lucentis) (Rader 2009).
PHAGE DISPLAY ANTIBODY LIBRARIES
Like natural antibody repertoires, antibody libraries are collections of millions to billions of different antibodies that collectively cover a large antigen binding space (Lerner 2006). Advances in antibody discovery and engineering have resulted in the development of robust techniques and tools for the generation and selection of antibody libraries, in particular display technologies that physically link phenotype (protein) and genotype (protein-encoding DNA or RNA). As such, they mimic and simplify B cells, which feature a more complex physical and functional linkage of exposed antibody and concealed antibody-encoding DNA and RNA. This universal concept is common to both cell-based (phage display, virus display, bacterial display, yeast display, and mammalian cell display) (McCafferty and Schofield 2015) and cell-free (ribosome display and mRNA display) (Amstutz et al. 2001) display technologies.
For more than three decades, phage display has been extensively used for the mining of naive, immune, and synthetic antibody repertoires from humans and a variety of nonhuman species (Barbas et al. 1991; Clackson et al. 1991; Lerner 2006; Winter 2019). Indeed, numerous mAbs in research and medicine have been discovered de novo or evolved in vitro by phage display, including what has now become the most profitable drug in history, adalimumab (Humira), an anti-TNFα mAb for the treatment of inflammatory and autoimmune diseases (Rome and Kesselheim 2023).
The main ingredients of phage display (Smith 1985) are filamentous phage particles that are derived from filamentous phage that belong to the genus Inovirus and infect Gram-negative bacteria, including E. coli. Filamentous phage particles used for phage display are elongated protein tubes that (1) “display” a peptide or protein of interest on the outside, (2) harbor the DNA encoding the peptide or protein of interest on the inside, and (3) retain the ability to infect E. coli. Importantly, each particle is designed to only display and encode a single peptide or protein. Amino acid sequence diversification of the peptide or protein of interest is presented by billions to trillions of particles (with a typical titer of 1012/mL), which collectively constitute a library. Using the displayed peptide or protein, the library can be stringently selected for or against binding to a small or large molecule, a cell or tissue, or other natural or synthetic objects of interest. Because the DNA encoding the selected peptide or protein is selected concomitantly, and the linkage of phenotype and genotype is stable, selected pools of particles can be reamplified by infection of host bacterial cells and subjected to several rounds of selection. This process is also known as affinity selection or, in laboratory jargon, panning (Smith 2023).
The generation and selection of antibody libraries typically use phagemid-based phage display systems that require three components; namely, phagemid library, host bacterial cells, and helper phage. A detailed review of phagemid-based phage display at the conceptual, functional, and molecular level can be found in this collection (see Overview: The pComb3 Phagemid Family of Phage Display Vectors [Rader 2024]). Briefly, phagemids are plasmids that contain the origin of replication and packaging signal of filamentous phage. After E. coli transformation, phagemids are replicated as double-stranded DNA (dsDNA), analogous to plasmids. When phagemid-transformed E. coli are superinfected with helper phage, however, phagemids are replicated as single-stranded DNA (ssDNA), for packaging into filamentous phage particles. Helper phage, which are wild-type filamentous phage particles with a debilitated packaging signal, provide all filamentous phage proteins and ssDNA required to assemble infectious filamentous phage particles. Converting a phagemid library to a phage library thus requires the efficient transformation of host bacterial cells, typically done by electroporation, followed by their superinfection with helper phage. The reason that phagemid-based phage display is the method of choice for antibody libraries is the use of minor coat protein pIII as the physical linkage of antibody and filamentous phage particle. Because pIII—of which five copies are presented in the spiky cap of the filamentous phage particle—is required for E. coli infection, recombinant fusion of an antibody fragment (such as a Fab) interferes with the ability to propagate the phage library. The phagemid/helper phage partition solves this problem by using the phagemid to encode a recombinant fusion of Fab and the C-terminal fragment of pIII (ΔpIII), and the helper phage to encode wild-type pIII. Phagemid-transformed E. coli superinfected with helper phage produce infectious filamentous phage particles that display zero to one Fab-ΔpIII copies and four to five wild-type pIII copies and harbor the phagemid ssDNA encoding the Fab-ΔpIII recombinant fusion (Fig. 1B). The excess of wild-type pIII, however, comes at the price of an excess of “bald” phage, which have zero antibody-ΔpIII copies, but allows monovalent display of the Fab, enabling affinity-driven as opposed to avidity-driven selections. Monoclonality is another important feature of phagemid-based phage display. Only one phagemid clone is replicated and packaged in the host bacterial cell, and only one filamentous phage particle can infect it. This ensures that the filamentous phage particles pair cognate phenotype (a Fab) and genotype (a phagemid clone encoding that identical Fab). As such, a Fab library is presented as a large collection of filamentous phage particles that allow the effective propagation, selection, and identification of Fab.
Depending on the origin of the Fab library, it can cover a large antigen binding space that can be selected for particular properties, such as high affinity [high association rate constant (kon), low dissociation rate constant (koff), or both], high specificity, species cross-reactivity, pH dependency, and high stability. Filamentous phage particles are assembled in the periplasm of E. coli, the space between the inner and outer membranes, which provides an oxidizing environment needed for Ig domains to fold into disulfide bridge-stabilized β sandwiches.
As mentioned above, the use of phagemids as phage display vectors enabled the expansion of phage display from peptide to protein libraries. Phagemid pComb3 was among the first phage display vectors used for the generation and selection of antibody libraries in the 50-kDa Fab format (Barbas et al. 1991). Smaller antibody fragments, including the 25-kDa scFv format, which connects VL and VH through a polypeptide linker, and the 12.5-kDa single-variable domain format known as nanobody, are alternative phage display formats of antibody libraries. A review of the pComb3 phagemid family of phage display vectors, which includes pComb3 successors pComb3H, pComb3X, pC3C, and pC3Csort, is part of this collection (see Overview: The pComb3 Phagemid Family of Phage Display Vectors [Rader 2024]). Collectively, pComb3 phagemids have been widely used for the de novo discovery of mAbs from natural (naive or immune) and synthetic antibody repertoires, as well as for the in vitro evolution of mAbs for affinity and specificity maturation and humanization.
The generation of an antibody library from an antibody repertoire encompasses the generation of a phagemid library that can be converted to a phage library suitable for various selection campaigns. A phagemid library and a phage library ideally fully capture and represent the amino acid sequence diversity in the antibody repertoire. In practice, however, this is difficult to achieve for natural antibody repertoires, as the original VL and VH pairs are cloned independently and recombined randomly (Beerli and Rader 2010). In antibody libraries that are derived from antibody repertoires of hyperimmunized animals, which are shaped by few VL and VH clonotypes, original or virtually original VL and VH pairs are restored and selectable. Naive antibody repertoires, on the other hand, can be diversified by random recombination of VL and VH, creating new selectable specificities. Synthetic antibody repertoires are typically built on one or a few defined VL and VH scaffolds, with amino acid sequence diversification confined to the CDRs (Zhai et al. 2011; Nilvebrant and Sidhu 2018; Teixeira et al. 2021).
GENERATION OF PHAGE DISPLAY ANTIBODY LIBRARIES IN FAB FORMAT
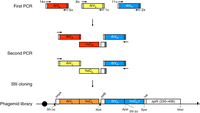
The first step in the generation of a phagemid library from a natural antibody repertoire is the preparation of total RNA from primary or secondary lymphoid organs (typically, bone marrow or spleen, respectively) or from peripheral blood mononuclear cells (PBMCs). The mRNA fraction is then reverse-transcribed to cDNA and subjected to PCR amplification using panels of sense and antisense primers that hybridize to the flanking sequences of VL- and VH-encoding cDNA. Suitable primer panels have been published for human antibodies, and also for antibodies from numerous other mammalian and avian species. To assemble Fab-encoding pComb3H and pComb3X cassettes (see Overview: The pComb3 Phagemid Family of Phage Display Vectors [Rader 2024]), pools of VL and VH cDNAs are first fused to CL and CH1 cDNA, respectively, via an overlap extension PCR and then fused to each other in a second overlap extension PCR. Following digestion with restriction enzyme SfiI and ligation into SfiI-cut pComb3H or pComb3X, electroporation of E. coli creates the phagemid library, with the number of independent transformants defining the maximum number of Fab clones. The Fab cassette, which is flanked by asymmetric SfiI (GGCCN_NNN^NGGCC) sites [shown as SfiI (a) and SfiI (b) in Fig. 2] and is under the control of a lacZ promoter/operon, equips VL-CL and VH-CH1 with N-terminal signal peptides for periplasmic transport in the host bacterial cell and results in the fusion of the C terminus of VH-CH1 with ΔpIII. The Fab-encoding pC3C and pC3Csort cassette is assembled by one three-fragment (VL-CL-VH) rather than by two two-fragment (VL-CL and VH-CH1) overlap extension PCRs (Hofer et al. 2007; see also Overview: The pComb3 Phagemid Family of Phage Display Vectors [Rader 2024]). This is achieved by moving the SfiI (b) site upstream of CH1. In this configuration, CH1 is encoded by the vector rather than the insert. This design is compatible with human as well as chimeric nonhuman/human Fab formats.
Assembly of chimeric rabbit/human “fragment antigen binding” (Fab) libraries in phagemid pC3C. Shown is a schematic of the workflow for the generation of a phagemid pC3C-based chimeric rabbit/human (rb/hu) Fab library, starting with a first PCR step in which rbVκ-, rbVλ-, and rbVH-encoding cDNAs are amplified from reverse-transcribed mRNA of a naive or immune rabbit antibody repertoire. Diverse primer pairs (14 × 5 for rbVκ, 8 × 1 for rbVλ, and 11 × 2 for rbVH) are used in 100 separate PCRs for each individual rabbit antibody repertoire. Subsequently, a second PCR step fuses the rbVκ and rbVH pools and the rbVλ and rbVH pools via a separately amplified invariable cDNA fragment that comprises huCκ and huCλ, respectively; a downstream ribosome binding site; and pelB. The flanking primers in the second PCR step also introduce the asymmetric SfiI (a) (5′-GGCCC_AGG^CGGCC-3′) and SfiI (b) (5′-GGCC_CCG^TCGGCC-3′) restriction sites, which are cut and ligated into the correspondingly cut phagemid pC3C. Electroporation of the ligation mixture into Escherichia coli completes the phagemid library, which typically comprises 108–1010 independent transformants. Addition of helper phage to the host bacterial cells converts the phagemid-encoding Fab library to a phage-encoding and displaying Fab library. In pC3C, a lac promoter (black circle) drives bicistronic ompA-VL-CL and pelB-VH-CH1-HA-ΔpIII expression cassettes, with ompA and pelB serving as signal peptides, HA (hemagglutinin) as a detection tag, and ΔpIII, which is a C-terminal segment of the minor coat protein pIII of filamentous phage. Shine–Dalgarno sequences are indicated as black triangles, and a transcriptional terminator sequence is depicted as a reverse black triangle. Annotated features of pC3C are discussed in detail elsewhere (see Overview: The pComb3 Phagemid Family of Phage Display Vectors [Rader 2024]).
Using bone marrow mononuclear cells (BMMCs) from healthy donors and PBMCs from patients, respectively, large naive and specialized immune human Fab-phage libraries based on pC3C have been generated and successfully selected (Kwong et al. 2008; Baskar et al. 2009). In addition, pC3C has been extensively used for the generation and selection of chimeric rabbit/human Fab-phage libraries to mine naive and immune rabbit antibody repertoires (Hofer et al. 2007; Peng et al. 2017; Weber et al. 2017). Thus far, three leads selected from pC3C-based chimeric rabbit/human Fab-phage libraries have been translated to clinical trials for cancer therapy (ClinicalTrials.gov IDs NCT02706392, NCT04441099, and NCT04877613). The workflow of building a chimeric rabbit/human Fab library in pC3C is shown in Figure 2. It is representative of the assembly of other Fab libraries that require different panels of sense and antisense primers for the PCR amplification of Vκ-, Vλ-, and VH-encoding cDNAs but use a set of identical primers for the subsequent three-fragment overlap extension PCR.
Irrespective of their origin, the generation of antibody libraries for phagemid-based phage display requires diligence with respect to protocols and reagents. Five accompanying protocols (see Protocol: Generation of Antibody Libraries for Phage Display: Human Fab Format [Peng and Rader 2024a]; Protocol: Generation of Antibody Libraries for Phage Display: Chimeric Rabbit/Human Fab Format [Peng and Rader 2024b] (Fig. 2); Protocol: Generation of Antibody Libraries for Phage Display: Preparation of Electrocompetent E. coli [Peng and Rader 2024c]; Protocol: Generation of Antibody Libraries for Phage Display: Preparation of Helper Phage [Peng and Rader 2024d]; Protocol: Generation of Antibody Libraries for Phage Display: Library Reamplification [Peng and Rader 2024e]) describe several methods that are broadly applicable and adaptable to the generation of antibody libraries from human and nonhuman naive, immune, and synthetic antibody repertoires.
SELECTION OF PHAGE DISPLAY ANTIBODY LIBRARIES IN FAB FORMAT
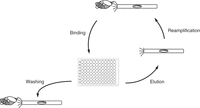
Once built, phage display antibody libraries can be subjected to selection using a variety of strategies tailored toward desired antigen binding properties (Ledsgaard et al. 2022). Antigens that are available as purified proteins are often immobilized on plastic surfaces, such as in wells of a 96-well ELISA plate (Fig. 3), followed by blocking with bovine serum albumin (BSA), incubation with the filamentous phage particles displaying the antibody library, and stringent washing. Remaining binders are eluted with trypsin or acidic buffer, added to host bacterial cells, and reamplified for another round of panning. As an alternative to immobilization on plastic surfaces, which can cause denaturation and epitope loss, antigens can be chemically or enzymatically conjugated to biotin and captured by streptavidin-coated magnetic beads. Filamentous phage particle binding to the beads via the antigen can then be isolated with a magnetic device. This method also allows selection of the antibody library using soluble rather than immobilized antigen if the biotinylated antigen and antibody library are mixed prior to adding streptavidin-coated magnetic beads. Gradually lowering the concentration of the soluble antigen over several rounds of panning favors the selection of high-affinity binders and is a common method in affinity maturation campaigns.
“Fragment antigen binding” (Fab)-phage library panning against immobilized antigen. Depicted is one round of panning that starts with adding a Fab-phage library to an antigen immobilized in one or more wells of a 96-well ELISA plate, for binding. Following stringent washing to remove nonspecific binders, specific binders are eluted using, for instance, trypsin (which removes the Fab phenotype but retains the Fab-encoding genotype) and then added to host bacterial cells for reamplification. Three to four rounds of panning are typically required to sufficiently enrich binders of high specificity and affinity from the Fab-phage library for identification by low-throughput screening. Screening methods using high-throughput sequencing only require one to two rounds of panning. Fab-phage libraries can also be selected against soluble antigen or on whole cells (see the text for details).
In cases where the antigen is not available as a purified protein but is targetable as an endogenously or ectopically expressed cell surface antigen, antibody libraries have been successfully selected on whole cells, typically mammalian cells (Alfaleh et al. 2017; Ledsgaard et al. 2022). Due to the presence of thousands of different cell surface antigens and the inherent stickiness of filamentous phage particles to mammalian cells, these panning campaigns are more challenging. However, they have been successfully applied to target-agnostic selections, enabling concerted antibody and antigen discovery. For example, we developed a modification of phagemid pC3C, named pC3Csort, which adds a C-terminal sortase A tag to the light chain of Fab (Wilson et al. 2018). This tag can be used for site-specifically introducing a biotin to filamentous phage particles that display Fab, while “bald” phage particles are not biotinylated. As such, specific binders can be efficiently separated from unspecific binders, facilitating target-defined and target-agnostic selections on whole cells (Wilson et al. 2018; Cyr et al. 2023).
One of the virtues of phage display is the ability to tailor the selection strategy toward unique binding properties that are so rare that they are difficult to find when mining antibody repertoires by screening rather than by selection campaigns. It allows identifying antibodies that can discriminate marginally varied amino acid sequences and conformations of highly homologous proteins by using one as bait and the other as decoy. Positive and negative selections can be done sequentially or simultaneously, and are also frequently applied to whole-cell panning (Alfaleh et al. 2017). When cross-reactivity to antigens (e.g., ortholog proteins from distinct species) is the objective, sequential positive selections can be applied, for instance, by alternating the bait over multiple rounds of panning. Elution of filamentous phage particles from immobilized or captured antigen can be tailored toward selecting, for instance, pH-, metal ion-, or small-molecule-dependent binding. In addition, selection campaigns can be designed to favor fast or slow association (high or low kon values) and fast or slow dissociation rate constants (high or low koff values) to diversify both thermodynamic and kinetic binding parameters of antibody–antigen interactions. Another selectable feature of broad utility is the stability of antibodies. In all, the ability of filamentous phage particles to tolerate substantial physical, chemical, and biological stress without losing integrity and infectivity has made highly stringent selection strategies possible.
To initially determine whether a selection campaign was successful, a phage ELISA designed to analyze the binding of phage pools collected after each round of panning is conducted. Subsequently, clones of interest are identified in a crude Fab ELISA and further analyzed by DNA fingerprinting and sequencing of the Fab-encoding phagemid cassette. Two accompanying protocols (see Protocol: Phage Display Selection of Antibody Libraries: Panning Procedures [Peng and Rader 2024f]; Protocol: Phage Display Selection of Antibody Libraries: Screening of Selected Binders [Peng and Rader 2024g]) provide step-by-step details for the selection of antibody libraries by phage display and their subsequent analysis.
Once Fabs of interest have been identified by this initial approach, they will need to be subjected to a set of rigorous analyses in order to determine their biophysical developability and biological suitability. These assays require pure Fabs of defined concentration. Their production is described in an accompanying protocol (see Protocol: Cloning, Expression, and Purification of Phage Display-Selected Fab for Biophysical and Biological Studies [Cyr et al. 2024]). Purified Fabs in milligram amounts enable advanced antigen binding analyses via, for instance, surface plasmon resonance, flow cytometry, X-ray crystallography, or cryogenic electron microscopy, which ultimately inform the success of phage display library generation and selection.
CLOSING REMARKS
The mining of human and nonhuman antibody repertoires by phage display has delivered mAbs of broad utility in research and medicine. A key aspect is the conversion of diverse and diversified antibody repertoires to antibody libraries that are suitable for stringent selection strategies. This requires the phage display of a monovalent antibody fragment that robustly and reliably represents mAbs, such as Fab. Indeed, the Fab fragment has been a favored format for phage display antibody libraries since their inception in 1991. Several generations of the pComb3 phagemid family of phage display vectors have been tailored for generating and selecting antibody libraries in Fab format, as described in eight accompanying protocols.
Footnotes
-
From the Advances in Phage Display collection, edited by Gregg J. Silverman, Christoph Rader, and Sachdev S. Sidhu