Analysis of Mouse Model Pathology: A Primer for Studying the Anatomic Pathology of Genetically Engineered Mice
- Center for Comparative Medicine and Center for Genomic Pathology, University of California, Davis, Davis, California 95616
Abstract
This primer of pathology is intended to introduce investigators to the structure (morphology) of cancer with an emphasis on genetically engineered mouse (GEM) models (GEMMs). We emphasize the necessity of using the entire biological context for the interpretation of anatomic pathology. Because the primary investigator is responsible for almost all of the information and procedures leading up to microscopic examination, they should also be responsible for documentation of experiments so that the microscopic interpretation can be rendered in context of the biology. The steps involved in this process are outlined, discussed, and illustrated. Because GEMMs are unique experimental subjects, some of the more common pitfalls are discussed. Many of these errors can be avoided with attention to detail and continuous quality assurance.
INTRODUCTION
Once an investigator has completed the genetic modifications of a new mouse model of cancer, the next step is to define the characteristics of that model. This article emphasizes the importance of context and combining molecular phenotype (function) with morphological phenotype (structure). Investigators using animal models should integrate the biology of gross and microscopic alterations with the associated molecular changes. However, it is important to consider basic biology! The pathology of cancer in genetically engineered mice (GEM) has been described in detail elsewhere (Cardiff et al. 2006c). We will concentrate on the responsibilities of the investigator for assuring that gross and microscopic pathology is placed into the context of the disease.
This “how to” article is designed to assist researchers in the planning and implementation of rigorous pathological analyses of samples (or specimens) from their research model. All members of the research team (principal investigator, faculty, fellow, staff, or student) are ultimately responsible for the quality of the pathological interpretation. Surveys reveal that about 95% of the interactions between mouse and scientists are carried out by “staff” (Schofield et al. 2009). Principle investigators, senior investigators, lead scientists, and pathologists rarely see the actual mouse and usually do not perform the data collection, gross observations, sampling, or histology. These experimental activities are usually provided by fellows, students, or staff, many of whom may have been trained by other (untrained) persons. One of the main goals of this article is to provide greater awareness of the need for training in observation and preparation of both gross and microscopic specimens. Technical hints are also provided and the necessity for careful observation of details is illustrated by specific examples from genetically engineered mouse models (GEMMs).
PATHOBIOLOGY OF CANCER IN GENETICALLY ENGINEERED MICE
What Is Pathology?
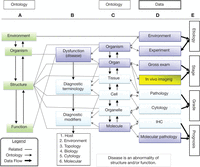
Pathology is the study of disease. Disease is an abnormality of structure and/or function. The interpretation of disease requires information from all levels of organization, from environment to molecule (Fig. 1). The study of disease structure is the unique domain of anatomical pathology that provides interpretation in the form of the “diagnosis.” The diagnosis involves putting all levels of information into the context of microscopic anatomy. Figure 1 illustrates the ontogeny of disease with emphasis on the roles played by various disciplines (Rosse and Mejino 2003; Cardiff et al. 2004). In the context of cancer, pathology is the study of the natural history, structure, and function of autonomous new growths in any multicellular organism. The goal of pathology is to understand the disease in the context of structure and function.
The ontogeny of disease. This ontology illustrates the complex relationships among structure, function, data sources, and diagnostic terminology (words used in diagnosis). It is based on the Rosse Foundational Anatomy ontology (Rosse and Mejino 2003; Cardiff et al. 2004). It shows the information required to understand microscopic pathology within the context of disease. The ontological structure provides an organ-based hierarchy with levels of anatomic organization to describe structure. The color in the boxes indicates the sources of the information: investigators (violet), imagers (yellow), and pathologists (light blue). Column A emphasizes normal relationships between organism, structure, and function. Column B demonstrates the relationship between the levels of organization and diagnostic nomenclature. Information is gathered from many levels of structure and function (column C). The pathologist's interpretation of the structure integrates all of the clinical, functional, and structural information (column D). Column E indicates the level of information used to decide the different clinical features of tumors in human medicine. The connecting lines indicate the relationships and flow of information (data) that provide context for the “diagnosis.” The diagnosis is primarily descriptive and adheres to standard terminology. However, as pathologists integrate more sophisticated data sources, the data must be recorded as “diagnostic modifiers.” If pathologists and scientists will adhere to this, or similar, ontology, computers and informatics will enable the scientific community to organize their data sets and recognize the relationships (“map”) across species. Investigators and pathologists will need to recognize and understand their roles in the organizational scheme to share data sources. This is our challenge and our responsibility.
All cancer investigators are students of pathology, in the sense that we all study disease. As a “para”-pathologist, you should be familiar with the basic vocabulary of disease. Table 1 provides a list of these general disease processes. Those who study the molecular biology of pathology are termed “molecular pathologists.” The discipline that is concerned with the structure of disease is termed “anatomic pathology.” Professional anatomic pathologists have unique training in, and understanding of, the microscopic anatomy of cancer. The uninitiated erroneously consider pathology as solely the study of microscopic anatomy, but the pathologist is an integrative biologist whose job is to place structural alterations into the context of the disease (Fig. 1). With proper information about the background of the research animal (context and function), the anatomic pathologist is capable of looking at a slide, which is a lesion fixed in time, to surmise its biological history. This capability amazes some observers and is developed from an in-depth study of the natural history of the disease.
General disease processes
The pathologist can integrate the individual sample back into the context of the dynamic natural history of the disease (Fig. 1). In the mouse arena, you and your team are the ultimate source of information about the pathobiological context of your mouse model. In many cases, it is the staff (rather than the principal investigator) who observe the live mouse, perform the necropsy, and take the initial steps in preserving and processing the samples (Schofield et al. 2009). As a result, it is incumbent upon the investigator and their staff to pinpoint areas of the gross anatomy and be able to adequately describe the key characteristics of each lesion, as well as to know how to document the disease, how to sample the specimen, and how to prepare it properly. The purpose of this article is to provide you with the basic guidelines for this task. The guidelines will be presented in the context of the pathology of cancer.
Pathology of Cancer
A neoplasia is an autonomous new growth (a tumor). A tumor is a physical (structural) entity that is identified as a focal mass in the animal (Figs. 2 and 3). Although the origins of neoplasia may be in somatic cell mutation(s) that drive massive alterations in the molecular and cellular biology of afflicted cells, neoplasia is a structural abnormality that can only be understood in the context of the whole animal. As intimated above, this morphology (structure) is the unique province of the discipline of anatomic pathology.
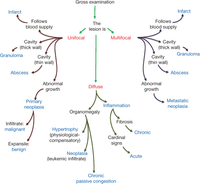
Gross specimen logic tree. Gross lesions can be divided into three types: unifocal, multifocal, and diffuse. In turn, unifocal and multifocal can be further divided into those that either follow the blood supply (infarct) or have a cavity with thick wall (granuloma), a thin wall (abscess), or an abnormal growth pattern (neoplasia). A diffuse disease pattern can result in organomegaly (hypertrophy, neoplasia, or chronic passive congestion) or from inflammation (chronic or acute). The diagnosis will be made/confirmed following microscopic examination (Fig. 3).
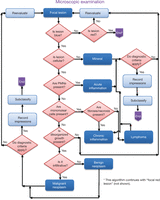
Examining a microscopic slide. This algorithm is designed to show the thought processes used by pathologists when examining microscopic slides. It will not make everyone in the laboratory eligible to take pathology board examinations, but we recommend that you follow the outline systematically. Step one: Look at a hematoxylin and eosin (H&E) stained slide with the naked eye. Try to orient yourself. Is it homogeneous in color? Is it mostly red? Is it mostly blue? Are different areas visible, such as an area that is pale and pink containing a mass of dark blue staining cells? Next, look at the focal mass with the microscope. Step two: Begin scanning the entire slide using the lowest magnification objective. Unless you are viewing hematology slides or slides containing microorganisms, oil immersion is never necessary. Identify the normal tissue and examine the interface between the normal and the abnormal mass (the lesion). As necessary, increase the magnification by changing objectives. Now, follow the steps indicated by the algorithm. Identify the cells that make up the mass. Clearly, a user needs to be able to identify PMNs (polymorphonuclear neutrophils), mononuclear cells, and disorganized growth. If not, a course in histology is recommended.
The study of the pathobiology of cancer should begin with an understanding of the stages of neoplastic progression: precancer, cancer, and metastasis. In principle, you should consider where the cancer starts, how it grows, and where it goes. To communicate these data elements with your colleagues and, specifically, the pathologist, it is important to properly collect and record the morphological and biological history of the disease. This information will lead to informed microscopic examination that confirms the gross findings and leads to accurate interpretations.
The critical background information in murine pathology includes husbandry, genetic background, gender, age, and experimental manipulations. In females, records of the parity and status of the estrous cycle may be critical. Environmental factors such as cage conditions and chow ingredients are important. If records are not meticulously kept, critical insights into disease can be overlooked or misinterpreted. For example, subtle changes in standard chow can lead to dramatic changes in tumor kinetics (Yang et al. 2003; Liu et al. 2005). In one case, moving immunologically impaired animals to a new vivarium resulted in changes in intestinal flora, opportunistic infections, and proliferative lesions in the gut (Borges et al. 2005). Without the environmental context, tumors can be misinterpreted (Fig. 4).
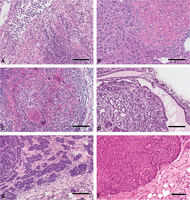
Pitfalls illustrated. These images illustrate a focal intestinal mass caused by roundworms (A,B), liver with EMH (C) and lymphoma (D), and an adenosquamous carcinoma before proper processing (E) and after reprocessing using appropriate techniques (F). The intestinal mass is shown as a screen capture of whole-slide imaging (WSI) viewed with the Aperio ImageScope web browser. (A) The low-power view with a large tumor mass bulging into the lumen. Scale bar, 4 mm. This was originally mistaken as an intestinal adenocarcinoma. However, close scrutiny revealed that the mass was composed of macrophages surrounding roundworms. Their hard, chitinous exoskeletons are easily visualized as boat-shaped blue profiles at higher magnification (B). Scale bar, 50 µm. The livers of mice frequently exhibit extra-medullary hematopoiesis (EMH) that differs from lymphomatous infiltrates in the size and nature of the nest and composition of the cells within the nest. EMHs tend to be smaller clusters with a mixture of cell types (C). Scale bar, 50 µm. Lymphomatous infiltrates present with more irregular profiles and a monomorphous cell population (D). Scale bar, 50 µm. The inadequately prepared adenosquamous carcinoma was poorly dehydrated and embedded in high-temperature paraffin (E), which proved impossible to cut thin without fracturing the tissue. Scale bar, 100 µm. The tumor was removed from the paraffin block and rehydrated and reprocessed. As a result, the reprocessed tissue could be cut at 5 µm in thickness and the fine details adequately viewed (F). Scale bar, 100 µm. To view the corresponding WSI for this figure, see the URLs provided in Table 3.
Most nonhematogenous neoplasms can usually be identified as a focal mass. The focal area may be completely confined to the epithelium but usually it will stand out as an atypical focus, both grossly and microscopically. The element of cytological atypia is the pathognomonic feature and requires microscopic confirmation. A neoplasm can be an entirely benign, indolent mass. Or it can be malignant with local invasion and/or metastases. The ultimate outcome is the metastatic spread of the neoplasm to distant organs. Because the investigator has access to all of this information, he/she should be responsible for knowing what to look for, how to look for it, and how to prepare it for the pathologist.
Hematogenous neoplasms are generally diffuse infiltrates that lead to enlarged organs (organomegaly) (Fig. 2). In mice, lymphomas and leukemias are recognized by enlarged lymph nodes, thymus, spleen, and/or liver. The murine spleen is a hematopoietic organ and a myeloid factory that readily responds to infection and other stresses. Therefore, splenomegaly will occur with cancer such as lymphoma, but also during almost any infection.
The reasons for careful observation and documentation can be illustrated by several examples. Mouse strains have different inborn, or spontaneous, disease patterns. For example, many strains spontaneously develop leukemias (Fredrickson and Harris 2000). Others, such as strains A and FVB, are very prone to bronchioalveolar adenomas, which are frequently misinterpreted as pulmonary metastases (Rehm et al. 1994; Mahler et al. 1996). Retired breeders of the FVB strain spontaneously develop diffuse mammary hyperplasia and pituitary adenomas (prolactinomas) (Mahler et al. 1996; Nieto et al. 2003; Radaelli et al. 2009). Afflicted FVB mice also have a higher incidence of mammary tumors (Radaelli et al. 2009). Some strains of immunodeficient mice are prone to osteogenic sarcomas, and all immunodeficient strains are susceptible to infectious diseases (Kavirayani and Foreman 2010). With accurate observation and detailed documentation, the critical differences between the diseases created by genetic modification and those that are spontaneous can be identified and understood.
Gross and Microscopic Tissue Examination
The gross examination begins with observation of the live animal and ends with necropsy. The behavior and general condition of the animal can indicate a disease state. Necropsy techniques are provided in Protocol: Limited Mouse Necropsy (Cardiff et al. 2014a) and have been described and documented online (http://tvmouse.compmed.ucdavis.edu and http://eulep.pdn.cam.ac.uk/Necropsy_of_the_Mouse/printable.php). In addition, a detailed atlas of normal mouse and human anatomy and histology has recently been published that includes an extensive illustrated section on necropsy (Knoblaugh et al. 2012; Treuting and Dintzis et al. 2012). One point that needs emphasis is the collection and examination of all tissues. This section will concentrate on the gross and microscopic characteristics of cancer in mice and the differential diagnosis of tumors (Figs. 2 and 3, Tables 1 and 2).
Microscopic key features: Focal lesions
As stated, neoplastic epithelial diseases are focal, space-occupying masses. This is their key gross feature. However, not all focal masses are malignant tumors. Abscesses, infarcts, granulomas, and benign neoplasms also present as focal masses (Figs. 2 and 5). Careful observation can sometimes distinguish between gross features of malignant neoplasms and other focal lesions. Figure 2 shows the logic tree that pathologists use to distinguish among various gross focal lesions. Malignancy is generally invasive and attached to adjacent tissue. A specific warning here: Many malignant tumors in mice have expansile, pushing borders and do not adhere tightly to adjacent tissue, but will still metastasize (Cardiff et al. 2006c) (Fig. 5).
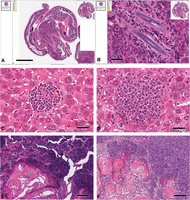
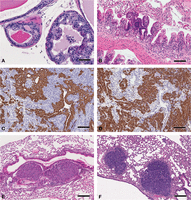
Pathologic processes. These images of H&E stained slides show the key microscopic features of the major processes associated with focal lesions. A (abscess) shows the center of an abscess with liquefactive necrosis and collections of polymorphonuclear leukocytes (PMNs). Note the loss of tissue detail. Scale bar, 100 µm. B (infarct) shows a hepatic infarct with coagulation necrosis. Note the bright pink cytoplasm with loss of nuclei. Scale bar, 100 µm. C (granuloma) shows a granuloma with an infiltrate of predominantly monocytic cells. Note the numerous multinucleated giant cells. Scale bar, 100 µm. D (benign neoplasm) shows a mammary adenomyoepithelioma within a duct-like space. Note the well-differentiated glands with a spindle cell stroma. Scale bar, 200 µm. E (malignant neoplasm) shows a Myc-induced (Tm(cMyc)) adenocarcinoma with poorly differentiated glands invading through a dense fibrous stroma. Scale bar, 100 µm. Contrast this invasive growth pattern with the expansile margin of the tumor in F. F (malignant neoplasm) shows the solid nodular profile of a Tg(MMTV-cNeu)-induced tumor with a peripheral palisade of basal cells and an expansile margin. Although not invasive, these expansile tumors can result in tumor emboli (see Fig. 6). Scale bar, 50 µm. To view the corresponding WSI for this figure, see the URLs provided in Table 3.
Gross differential diagnosis of tumors is described in Figure 2. In contrast to malignant tumors, abscesses are cystic with thin walls and are filled with a yellowish liquid (pus). Some chronic abscesses have a fibrotic host response and may adhere to adjacent tissues. The purulent fluid of an abscess can mimic the necrotic center of a neoplasm but is generally less viscous. Granulomas are the focal result of granulomatous inflammation, which is a type of chronic inflammation dominated by monocytes and their derivatives such as multinucleated giant cells. Mice rarely form grossly visible granulomas. Infarcts are relatively rare in mice but can be confusing. They are the result of vascular occlusion and typically follow the blood supply. Therefore, one needs to know the vascular patterns of the organ under study (Fig. 5).
The gross observations, however, are just “impressions” and the diagnosis must be confirmed by microscopic examination. The logic used to distinguish between the basic lesions upon microscopic examination is represented in algorithmic format with key features in Figure 3. All too frequently, morphology is sacrificed for molecular studies. Without a histological sample of the tissue to verify that it is a tumor, the data on an expression microarray can be misinterpreted and misleading.
Another limitation to gross-only diagnoses is that some mouse models have tumors with different phenotypes (see discussion below). Even worse, this heterogeneity can occur within the same neoplastic mass. Once again, the gross impressions need to be confirmed microscopically to avoid misinterpretation of the molecular data.
Natural History of Cancer and How to Identify Precancers
Carcinomas arise from precursor lesions, which are variously referred to as carcinoma in situ, preneoplasms, precancers, and/or premalignancies (Cardiff and Borowsky 2010, 2011). All forms of epithelial precancers are characterized as focal areas of atypical cells with an increased risk of malignant transformation (Cardiff et al. 2006a). This implies that they are not yet composed of malignant cells but require another molecular hit. They can be observed at the subgross level, using a dissecting microscope, as small foci in the organ without evidence of invasion. Although difficult to see in solid organs, they can be found by experienced observers in the mammary gland, skin, lung, gut, and prostate. In human medicine, their biological potential is judged by their relationship to malignancy. The documentation may require years of observation and extensive epidemiological evidence to establish a relationship. This is essentially guilt by association; evidence is rarely obtained by direct observation because, in human medicine, the diagnostic tissue has been removed and processed. In the inbred mouse, however, experimental evidence can be obtained using orthotopic and ectopic transplantation (Cardiff and Borowsky 2010, 2011). This is known as “test by transplantation” (Cardiff and Borowsky 2010, 2011). Basically, if the focal lesion/tumor mass will not grow as a transplant in an ectopic site, it is not considered to be a malignant neoplasm. If it does grow in an orthotopic site where it eventually becomes a tumor, it is considered a precancer or a neoplasm with malignant potential.
Microscopically, a precancer is characterized by a focal area of dysplastic cells that is confined within the basement membrane or normal tissue limits (Fig. 6) (Cardiff et al. 2006a). The key features are the abnormal growth and dysplastic cytology (Fig. 6). Detailed descriptions of specific precancers can be found elsewhere. Fundamentally, these lesions will stand out from the background of normal tissue because their cells are darker staining (hyperchromatic) and are larger and more irregular (pleomorphic) than normal. In addition, normal tissue organization is disrupted. The nuclei are larger with a higher mitotic rate. Unfortunately, it is impossible to predict biological behavior based on cytological criteria (Medina 1996; Cardiff et al. 2006b). As with any pathological lesion, a differential diagnosis must be considered. Occasionally, a systemic condition can lead to widespread atypical hyperplasia. In these cases, such as some inflammatory bowel diseases, the entire epithelium might seem to be dysplastic but there are no focal lesions. Some focal lesions are the result of injury and do not progress to malignancy. The inflammatory nodules in postlactational mammary glands are examples of such lesions (Raafat et al. 2012).
Precancer, tumor heterogeneity, and metastasis. These are examples of precancer (A,B), tumor heterogeneity (C,D), and tumor emboli and colonization in the lung (E,F). (A) Low-grade PIN (prostate intraepithelial neoplasm) in a Tm(Pten−/−) male. Multilayered areas of atypical cells (center) stand out from the glands with normal cells (11 and 1 o'clock). Scale bar, 50 µm. (B) A small zone of hyperchromatic, dysplastic cells occupying a crypt in the small intestine of an APC-MIN mouse. Scale bar, 100 µm. (C,D) Serial sections from a transplanted tumor cell line derived from a Tm(Stat−/−) mammary tumor stained for CK19 (C) and CK5 (D). Scale bar (C,D), 100 µm. Note the branching vascular space at right center outlined by CK19-positive (C) but CK5-negative (D) cells. (E,F) Pulmonary involvement with tumor emboli within vascular spaces (E) that are inside the vessels. In contrast, the tumor metastasizes into the lung with colonization of the lung parenchyma (F). Scale bar (E,F), 100 µm. To view the corresponding WSI for this figure, see the URLs provided in Table 3.
Malignancy
Malignant neoplasia is life-threatening. In its early stages, the neoplasm invades the surrounding tissue or the vasculature (Fig. 6). The ultimate stage is its spread (metastasis) to distant sites. The very earliest stage involves microinvasive disease. The criteria for microinvasion include extension through the basement membrane by distinctively dysplastic cells and a host response to the invasive cells (Shappell et al. 2004). Unfortunately, most examples lack one or more of these criteria and even experienced pathologists have difficulty interpreting the histology. In these cases, the better part of valor is to recognize that criteria are not met so that the lesions are designated “suspicious” or “probable” for microinvasion (Shappell et al. 2004). This cautionary approach is particularly important in epithelial lesions of mice. The intestinal herniation phenomenon is one example in which specific knowledge of mouse anatomy is critical and caution is necessary. The mucosa of mice has a tendency to herniate through the muscular wall of most tubular organs, such as intestines. In inflammatory conditions of the bowel, reactive epithelium may bulge through the muscularis. The condition is reversible if the inflammation is treated and regresses. If the pathologist is not aware of this tendency, they can “overcall” microinvasion and cause a futile search for more convincing evidence of malignancy. The test by transplantation can provide the definitive experimental proof if the histopathology is equivocal (Cardiff and Borowsky 2010, 2011).
Metastases
Distant spread of the neoplasm (metastasis/es) is the endpoint of the neoplastic process. Here again, the mouse has a number of peculiarities that will be highlighted below. In contrast to most human cancers, slower growing tumors, not the highly proliferative tumors, are more likely to result in metastasis in mice. Also, in contrast to most human carcinomas, which primarily spread to local lymph nodes and to specific organs, the vast majority of mouse cancers metastasize to the lung through the hematogenous route and do not invade lymphatic channels (Fig. 6). With diligent searches, metastases may be found in regional lymph nodes, liver, and other organs. However, metastases to these other tissues tend to be rare in the mouse.
Another issue is the large proportion of metastatic tumors that arrive in the lung as tumor emboli (Fig. 6) (Cardiff et al. 2006c). Frequently, these emboli arrive carrying their own coating of endothelial cells (Oshima et al. 2004; Cardiff et al. 2006c). The tumor emboli can clog up the major pulmonary vessels without ever invading or colonizing the lung. As a result, experienced pathologists keep a record of different types of emboli. For example, it is conceivable that small clusters of cells arrive at the lung only to be trapped in the peripheral vasculature but do not invade through the vessel wall. They may also grow (enlarge) without colonizing the lung. A number of investigators have recorded a transient time period when tumor cells will be trapped in the lung or liver before they break loose and travel to colonize a secondary organ (Fidler and Poste 1982; Poste 1982; Poste and Nicolson 1983; Morris et al. 1994). Again, these types of tumor microemboli should be recognized and enumerated (Siegel et al. 2003). Most authorities insist that the diagnosis of pulmonary metastasis be reserved only for those tumors that colonize the lung parenchyma. Figure 6 illustrates tumor emboli and metastatic colonization in the lung.
Bronchioalveolar adenoma/carcinoma (BAC) (Rehm et al. 1994) is another entity that frequently causes confusion. These tumors are very common in older females of some strains, such as Strain A and FVB. Many uninitiated observers will misinterpret these lesions as metastatic. Most BAC are papillary or lepidinal and do not resemble the primary tumor. Further, they are associated with the bronchi and not the vasculature.
Since the mouse liver is a common site for extramedullary hematopoiesis (EMH), these foci are sometimes mistaken for metastases or diagnosed as lymphoma. On H&E staining, EMH foci can usually be identified by their small size and round, well-circumscribed profile. The dominant cell type is myeloid and its derivatives create a morphologically diverse, mixed cell population. In contrast, most lymphomas are monomorphous (single-cell type) and form larger, more irregular foci (Fig. 4). Sometimes, these lesions can be differentiated from each other by using immunohistochemistry to determine whether the cells are from the same or mixed lineages (Fig. 7).
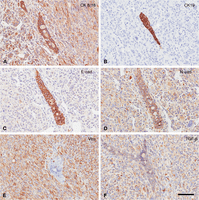
Immunohistochemistry. An illustration of the molecular heterogeneity of specific antigens using immunohistochemistry in an epithelial–mesenchymal transition (EMT) tumor from a Tm(cMyc) mammary gland. Note the tumor has a mixture of solid nests/glands and spindle cell elements that are typical of EMT phenotype tumors. Also note that both the epithelial nests/glands and spindle cells contain CK8/18 (A), N-cadherin (D), and TGF-β (F). The spindle cells contain vimentin (E) but not CK19 (B) and a relatively small amount of E-cadherin (C). E-cadherin and CK19 are primarily found in the epithelial component. The dual location of the intermediate filaments is considered diagnostic of EMT tumors. Scale bars, 100 µm. To view the corresponding WSI for this figure, see the URLs provided in Table 3.
Neoplastic Variation, Heterogeneity, and Reproducibility
The study of GEM pathobiology has led to the recognition of gene- and pathway-specific tumor phenotypes (Rosner et al. 2002) and oncogene addiction (Cardiff et al. 2011; Couto et al. 2012). Gene-related morphology reflects the molecular biology of gene addiction. These phenomena are consistent with our current concepts of tumor clonality. Further, the loss of the inciting oncogenic transgene is associated with a distinctive change in the tumor morphology. As tumor biologists, we need to be aware that even clonal tumors can be variable. This biological and morphological variation and heterogeneity of all tumors should be recognized, documented, and understood (Fig. 7). This is probably one of the most confounding aspects of tumor pathology. Although the pathologist can predict outcome on the basis of the current appearance of the neoplasm, the prediction is frustratingly imprecise. The goal of molecular pathology is to increase the prognostic and predictive precision of the diagnosis. It is envisioned that molecular clues will be more predictive than morphological clues. To some extent, these clues have been transforming oncology.
Science technology has become more sensitive and sophisticated. The search has intensified to find the key characteristics of malignant neoplasms that can be used to prevent, treat, and hopefully cure cancer. Huge national and international programs are dedicated to cancer genomics and biomarkers. In part, this search has been driven by the success of the reductionist approach of molecular biology and, in part, by the dazzling power and unexplored potential of current and emerging technologies. The stumbling block to the realization of this vision can be found in the amazing variation between and within neoplasms.
Some authorities argue that every tumor is different and each has infinite potential for variation with a complexity that can never be completely understood (Berman 2004). The clonal origin of most tumors would argue against this rather nihilistic hypothesis of infinite variation. The current enthusiasm for cancer stem cells also argues for a common origin (Damonte et al. 2007; Garbe et al. 2012). The issues are the speed at which subclones evolve and whether subclones retain the characteristics of the primary clone.
In support of the primacy of the original clone, one can cite the remarkable genotype-specific phenotypes of the strong initiating oncogenes used to create GEM (Rosner et al. 2002). Activation of most transgenic oncogenes results in very specific tumor morphology (phenotypes) (Cardiff et al. 2006c). An experienced comparative pathologist can recognize tumors associated with many genetic manipulations. For example, it is easy to recognize changes induced by Ras, Myc, Wnt, PI3 kinase, and ErbB2 activation; inactivation of Pten or p53 in the mammary gland; activation of Ras, pAkt, and SV40 in the prostate; and activation of Ras or T antigen in the lungs (Cardiff et al. 2006c). This phenomenon has been discussed under the heading of oncogene addiction and non-oncogene addiction (NOA) (Cardiff et al. 2011; Couto et al. 2012). These types of morphological observations reinforce the concepts of clonal origin and the potential for customized medicine. However optimistic, these concepts are limited when the variability and heterogeneity of malignant neoplasms are recognized.
In spite of the similarities emphasized in oncogene and NOA, each signature phenotype has variation between tumors and even within tumors in the same animal. For example, although Tg(Wnt) tumors may share certain features such as the retention of the myoepithelium, they may also have as many as five different tumor phenotypes in the same animal (Rosner et al. 2002). Myc-derived tumors have the same large, hyperchromatic blue cell phenotype but their tumors will vary in the details of their histological organization (Andrechek et al. 2009; Leung et al. 2011). The biological potential of these variations has not been studied nor have expression microarray studies been correlated with the microscopic tumor pathology.
The situation becomes more complex when discussing GEM tumors that have escaped oncogene addiction. These recurrent or persistent tumors generally have lost the expression of the activating oncogene. The most extensively documented histological studies of escape from addiction involved the phenomenon of epithelial-to-mesenchymal transition (EMT) (Cardiff 2010). The tumors that undergo EMT become increasingly less differentiated and end up as spindle cell tumors (Fig. 7) (Radaelli et al. 2009). They can be differentiated from sarcomas by the presence of intermediate filaments characteristic of both mesenchyme (vimentin) and epithelium (keratin) (Damonte et al. 2007). Interestingly, this EMT-type transition is accompanied by an increase in local invasion and loss of metastatic potential (Cardiff 2010). The EMT tumors, regardless of the initiating oncogene, seem to be a common final pathway for diverse oncogenes during neoplastic progression.
Mouse tumorigenesis seems to be particularly prone to EMT. The term EMT was not introduced until 1989, but, in retrospect, mouse mammary EMT has been observed from the very beginnings of experimental mouse pathology. It was originally attributed to transplantation artifact (Cardiff 2010). When in vitro culturing of tumors was developed, many of the cultured cells became spindle-shaped and the EMT phenomenon was thought to be a result of tissue culture. With the development of evidence for oncogene addiction, based largely on GEM, escape from oncogene addiction was found to be a result of EMT. Although EMT has been extensively studied in the context of the mammary gland, EMT occurs in the context of other organ systems, such as skin and prostate (Lacher et al. 2006; Ding et al. 2012).
The lesson to be learned when dealing with tumors of all types is that they are heterogeneous. The investigator cannot assume, because two tumors arose in a molecularly defined animal, that the phenotype or biological history is known. In addition, investigators need to be aware of transplantation and tissue culture and their effect upon tumor variability. Both are very valuable techniques, yet they have a profound effect upon the critical characteristics of a tumor.
TECHNICAL PREPARATION FOR STUDY OF GEM
The investigators and their pathologist need to know the details of the mouse model being used (Fig. 1). Presumably, the environment, experiment, gross examination, in vivo imaging, and molecular biology are all documented and made available to the pathologist. Ideally, the documentation is in a well-organized format in a database with controlled, searchable terminologies and common data elements (Cardiff et al. 2004) (Fig. 8). If any one of these elements is neglected, the analysis of the pathology may be incomplete. Complete documentation involves integration of diverse sources of information. In modern times, it means using electronic formats with the use of proper nomenclature and controlled vocabularies (Cardiff et al. 2004). The final form of documentation is the presentation for publication.
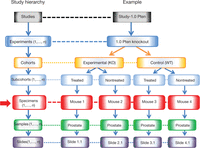
Study hierarchy. This figure depicts the application of the ontogeny for experimental design. The ontogeny, labeled Study hierarchy, follows Aristotelian rules. In most clinical settings, the organism, be it a mouse or human, is the subject (red arrow) and carries the universal identifier. The Specimen (red box) (or specimen) is the equivalent of Organism in Ontology C (violet box) of Figure 1. The Slides are the children of a Sample, the Samples are children of a Specimen, the Specimens are children of a Subcohort, the Subcohorts are children of a Cohort, the Cohorts are children of an Experiment, and the Experiments are children of a Study. In practice, Slides belong to an organ (Sample) (e.g., prostate) in an animal (Specimen) (e.g., Mouse 1, …, n), all Specimens belong to a Subcohort (treated or untreated), Subcohorts belong to a Cohort (Experimental or Control), Cohorts belong to an Experiment (e.g., Pten KO) and all Experiments belong to a Study (e.g., Pten). By using these simple rules, the investigator and computer can track the origin and distribution of multiple levels of information while maintaining order in relationship to each animal and their samples. This ontogeny is also consistent with the anatomic ontogeny in Figure 1.
Great concern and demand has been expressed for developing “standards” for anatomic pathology (histopathology). Most demands focus on the person interpreting the slides, the anatomic pathologist. However, as has been outlined above, pathologists are limited by the quality and quantity of the information about each mouse and the quality of the preparations presented to them (Fig. 4). As stated above, the pathological analysis begins with the person examining the mouse. In most institutions, the pathologist is a specimen's “last stop.” Therefore, the investigator has primary responsibility for the initial observations, descriptions, dissections, and processing of the specimen. The investigator is responsible for understanding the normal gross anatomy and identifying abnormalities. Thus, we have outlined the basic information the investigator should know about cancer and neoplastic progression. Next, we will deal with technical preparations (necropsy and tissue fixation) and choosing a pathologist.
The Necropsy
The investigator should be very familiar with murine gross anatomy and, specifically, the anatomy of the organ of interest. Reference guides are available for dissection of specific organs. The recent Comparative Anatomy and Histology: A Mouse and Human Atlas is a good place to start (Treuting and Dintzis 2012). If it is already known that the animal has a tumor, plan the dissection in advance and be prepared to triage portions of the tumor to appropriate sources using the proper fixative for the test being performed. General instructions for sampling and preparing murine organs have been published (Ruehl-Fehlert et al. 2003; Kittel et al. 2004; Morawietz et al. 2004). Refer also to Protocol: Limited Mouse Necropsy (Cardiff et al. 2014a).
Fixation
Fixation stops postmortem changes that degrade tissue and allows optimal preservation of morphologic and cytological detail as well as nucleic acid integrity. Following death, tissues soon undergo autolysis, and if organisms from the gastrointestinal, urinary, or respiratory tracts are present, their colonization can soon cause putrefaction. Placing the tissue into a fixative stops these postmortem changes. See Protocol: Mouse Tissue Fixation (Cardiff et al. 2014b) for a basic tissue fixation procedure and guidance on choosing an appropriate fixative, the timing and duration of fixation, sample storage, and quality issues.
Handling Tissues after Fixation
Fixed tissues are transported to the appropriate facility for the procedures required by the experiment. That facility can be your own or an “outside” laboratory. Regardless, if you are primarily interested in genetic studies, there should always be a portion of the tissue set aside for H&E staining. This will provide an answer to “What was the tissue that was evaluated?” If other, esoteric evaluations are requested, it is important to check with the histology laboratory performing the procedures to be certain all requirements are met. Needless to say, this should be done before the necropsy. Protocols for routine H&E staining and also for immunohistochemical staining are given in Protocol: Manual Hematoxylin and Eosin Staining of Mouse Tissue Sections (Cardiff et al. 2014c) and Protocol: Manual Immunohistochemistry Staining of Mouse Tissues Using the Avidin–Biotin Complex (ABC) Technique (Cardiff et al. 2014d).
Interpretation and Choosing a Pathologist
To obtain proper interpretation of your specimens, a pathologist (preferably one with experience in GEM pathology) needs to view your records and slides. The microscopic examination is something anatomic pathologists actually do control. You should approach the pathologist with realistic expectations. The wise investigator identifies and works with their pathologist before the project starts. Seeking advice after the project is completed is too late for most remedial action. The pathologist's job is to integrate the microscopic observations with all other sources of information about the disease in the animal. The diagnostic interpretation is provided in context.
What attributes are required of a pathologist? Basically, two kinds of pathologists exist: diagnostic and discovery pathologists. The diagnostic pathologist will verify that you do, or do not, have a tumor and attach a name (diagnosis) to your slide and move on to the next slide. This superficial behavior is currently used for high-throughput clinical diagnoses and, in investigative pathology, has led to any number of errors in the literature (Fig. 4). Pseudopathology is particularly rampant when the pathologist is not trained in comparative pathology. Examples are numerous and include mistaking neuroendocrine tumors for adenocarcinoma and mistaking the rodent preputial glands for teratomas and mouse nipples for “premalignant” papillomas (Barthold et al. 2007; Cardiff et al. 2008; Ince et al. 2008).
If the investigator is looking solely for a “name” (diagnosis), satisfaction will be achieved if the name fits their bias. If it does not, some investigators will try to find another pathologist who tells them what they want to hear. This behavior is called “shopping for a pathologist.” Or an investigator can ignore the pathologist and apply his or her own preconceived nomenclature. This is called “do-it-yourself” (DIY) pathology (Ince et al. 2008). These types of behavior end in well-documented scientific gaffes (Couto and Cardiff 2008; Ince et al. 2008).
The second type of anatomic pathologist is the one who should be sought for your unique study: the discovery pathologist. Discovery pathologists will not only name the disease but will want to put the disease into biological context. They are interested in the study of disease and want to use your mice to learn more. Our experience has been that most true discovery pathologists are eager to learn about the biological process in the experimental mice they are studying. However, it is most beneficial if the investigator engages them early in the process and not use them as the final step in the experiment. Because the mouse models are intended for the study of human disease, the investigator should choose comparative pathologists with sufficient knowledge of both species to avoid errors. Medically trained pathologists should be able to recognize the spontaneous tumors in your mouse strain and compare them with the gene-induced tumors. Ideally, they should also be “genomic pathologists” who are well-versed in modern molecular biology and nomenclature. Most important, the pathologist should engage the investigator in the learning experience.
Unfortunately, trained comparative pathologists are relatively rare. Various training programs are being devised to fill this need but a shortage remains (Barthold et al. 2007; Cardiff et al. 2008; Warren et al. 2009). Workshop training courses are available (Sundberg et al. 2012). A group of comparative pathologists has banded together under the banner of the Center for Genomic Pathology (http://ctrgenpath.net) (Cardiff et al. 2008; Couto and Cardiff 2008). They can either provide the investigator with assistance or direct you to the appropriate resources. The Center for Genomic Pathology has developed online courses aimed at graduate students, fellows, and new investigators. In cooperation with UC Davis Extension, CGP has seminar-type interactive programs for Principle Investigators (http://www.extensiondlc.net/).
HANDLING EXPERIMENTAL PATHOLOGY DATA
Data Organization and Integration
Little has been written to guide the laboratory documentation of experimental studies of GEMMs. The documentation is generally considered the purview of the individual laboratory and, as it should be, the responsibility of the principle investigator. Although this all seems logical, in an unpublished survey carried out in 2005 for the National Cancer Institute's Mouse Models of Human Cancers Consortium, the principle investigators of five prestigious laboratories were asked to show their documentation for recent publications. None of the five were able to show the primary data for the published experiments. The primary data were said to exist in a collaborator's laboratory or in the now-missing laptop of a former fellow. None could show evidence that technical data provided by another university actually belonged to a specific mouse. The best the investigators could do was to show PowerPoint presentations, which were representations of the data but not the actual data.
Moreover, the scientific community has expanded. Core laboratories are called on to provide specific data sets. Collaborative studies are encouraged. In modern times, data need to be collected in electronic format and coordinated in a common, accessible server. The principle investigator must take responsibility for the collection, integration, and integrity of the data. The following sections will provide some suggestions of how the neophyte can conceptualize, organize, and document their data.
A logical hierarchy for recording pathology data has been developed and successfully applied (Fig. 8). The hierarchy organizes data according to Study, Experiments, Cohorts, Specimens, and Slides. In this experimental hierarchy, each Slide belongs to a Sample, each Sample belongs to an animal (Specimen), each animal belongs to a Cohort and subcohort, each Cohort belongs to an Experiment, and each Experiment belongs to a Study. This hierarchy can be divided into subcategories. For example, Experiments frequently involve experimental and control animals. Each is considered a separate group or Cohort. The experimental groups may contain multiple Cohorts and each Cohort multiple subcohorts (such as dosage or other experimental manipulation).
When data concerning a given animal arrive from another laboratory, the data need to be identified or tagged by the identifier from each laboratory. For the pathologist, the animal's samples can be divided into separate blocks for processing. Further, different slides from each block may be stained using different techniques. A common identifier must be used for all of the slides, samples, and specimens.
We use a laboratory accession number that identifies the animal to track all data. The computer also records the animal ID provided by the investigator which links the slide to the Specimen, Cohort, Experiment, and Study. When the slide is recorded by whole slide image (WSI; see below) or as a “point-of-interest” still image, a standard image file format is used that includes accession number-block-photograph-stain-magnification-camera-photographer-subject. For example, file name EX12-0420-8-A-HE-x20-Ax1-RJM-MIN denotes image A from block 8, on a H&E-stained slide from an animal with accession number EX12-0420, captured with the 20× objective on a Zeiss AxioCam by operator RJM, showing an image of a mammary intraepithelial neoplasm (MIN).
We have developed and continue to maintain a database that is customized with the appropriate hierarchies and data fields. Some fields use a required (controlled) vocabulary but others permit free text. Because we have many submitters, the entries need to be audited throughout the experiment to ensure data integrity. The auditors must address whether the data are searchable. To the extent that we can control entries from multiple sources, the data are searchable. We have 22 years of data from well over 500 laboratories and submitters. This article is a demonstration of the power of such archives: Almost all of the examples were identified by searches of our database.
Nomenclature: Understanding “Pathology-Speak”
In this modern era of electronic capture, storage, and retrieval, controlled vocabularies become absolutely essential. Considerable effort has been given to controlled vocabularies and synoptic reporting in health care and in experimental pathology (Cardiff et al. 2004; Schofield et al. 2009; Schofield et al. 2010a,b, 2011; Gaudet et al. 2011; Hoehndorf et al. 2011). Ontologies of disease are useful in this exercise (Fig. 1). However, most research laboratories do not have immediate access to these systems. As research becomes increasingly more complex and sophisticated, the comparative pathology of all species, let alone human and mouse, will inform us. Ultimately, the investigators who can relate their models to the rest of comparative pathobiology will have the most impact. Therefore, it is incumbent for the neophyte to learn proper nomenclature and use synoptic reporting.
A survey of the current terminologies will reveal that some discrepancies still exist between human and mouse nomenclature (Rudmann et al. 2012). The major discrepancies involve the current separation between structure and function. The standard mouse nomenclatures are not supported by molecular data. Thus, they are based almost completely on descriptive morphology. The human classifications are almost completely based on gene expression profiles with relatively few attempts to match to the morphology. The genetically modified mouse is the one model system in which structure and function have been matched (Cardiff et al. 2006; Rudmann et al. 2012).
Nonetheless, the pathology community has worked hard over the past two decades to provide controlled vocabularies and standard nomenclatures (Schofield et al. 2010a,b). A huge international cooperative sponsored by the Society of Toxicological Pathology periodically reviews, updates and “harmonizes” diagnostic nomenclature (Renne et al. 2009; Thoolen et al. 2010; Rudmann et al. 2012). Numerous consensus reports and harmonized vocabularies have been published to assure the scientific community that we are using similar, if not the same, vocabulary (Cardiff et al. 2000; Kogan et al. 2002; Nikitin et al. 2004). Comparative anatomy monographs and atlases are also available as references (Holland 2004; Bissell et al. 2011; Treuting and Dintzis 2012).
At the laboratory level, a difficult problem is nomenclature, because of the human tendency to personalize data elements. The worst offenders choose to enter their data independently, while ignoring the rules enforced by the chosen hierarchy. Because we work with people around the world, we have encountered different levels of data organization as well as complete lack of data organization. Inside the individual laboratory one finds varying degrees of compliance. As a result, someone from each laboratory should be responsible for vocabulary compliance and data integrity. Failure to meet this responsibility can result in the loss of valuable data.
Presenting the Images
Almost all pathology is now represented in publications as microscopic images, and the legends use diagnostic terminology rather than descriptions. Unfortunately, this mode has a number of shortcomings. Any experienced pathologist can determine the relative knowledge of the person capturing the images with a simple glance. The photographer should know the rudiments of proper microscope adjustment. The most common error is the peripheral vignette in the image. When present, it is obvious the photographer does not know microscopy. Anyone unfamiliar with Köhler illumination should learn to use it for optimum adjustment of the microscope.
Naïve investigators who have been misled by less-experienced pathologists have presented us with images labeled as “teratomas” that were in fact preputial glands (Ince et al. 2008), “premalignant papillomas” of the skin that were in fact nipples (Coste et al. 2007), “adenocarcinomas of the prostate” that were in fact neuroendocrine tumors (Couto and Cardiff 2008), “adenomas” with invasive margins, “colonic cancers” that were, in fact, submucosal granulomas associated with parasitic infestations (Borges et al. 2005), and the list goes on. The point here is that the images were not only misleading, but the diagnosis was incorrect. The critical consideration in all of these examples is whether or not the images showed the appropriate field in their small thumbnail images.
Often manuscripts present multiple thumbnail images that seldom are informative. This is known as “postage stamp” pathology. These images are highly selective and may not necessarily represent the overall process. Second, the images are too small for any legitimate pathologist to make a diagnosis or verify the written interpretations in the text. Unfortunately, the microscopic images found in this article are good examples of this frustrating practice (Figs. 4, 6, and 7). When the images are used to represent immunohistochemistry, one can generally only determine whether the cells are brown or not; they rarely contain positive and negative controls that indicate sensitivity and specificity of the stain. Pathologists who are experienced in the field or have participated in Consensus Workshops frequently find that the published images do not accurately reflect what is actually on the corresponding slides.
Whole-Slide Imaging (WSI)
Many, if not most, institutions now have instruments that can digitize entire microscopic slides at high resolution so that they can be viewed at almost any magnification at any point on the slide, anywhere in the world. These are invaluable instruments for storing and annotating microscopic images. They are now used on a daily basis for education and conferencing. They provide the viewer with a comprehensive view of the entire slide from remote locations. WSI should also be used to represent pathology in manuscripts. At the very least, these images should be made available for the reviewers, who could prevent the egregious errors that currently plague publications. However, publishers have been very slow to adopt WSI and have yet to come up with a reasonable business plan. Until the time comes that publishers can profit from the current technology, they will continue to publish “faux” pathology. Beware of what you see in the hard-copy publication! Although we cannot show you the WSI directly, Table 3 provides URLs to WSI of Figures 4–7.
Links to whole slide images (WSI) for selected figures
SUMMARY AND CONCLUSION
This primer of pathology emphasizes the necessity of using biological context for the interpretation of anatomic pathology. The steps involved are outlined, discussed, and illustrated. Some of the pitfalls are discussed and illustrated. These errors can be avoided with attention to detail and continuous quality assurance. This primer is not a comprehensive review of pathology but should stimulate you to increase your awareness and knowledge of what pathologists can do and how they do it.
ACKNOWLEDGMENTS
The research described here was supported by grants U01 CA141582, U01 CA141541 and U01 CA105490-01 from the National Cancer Institute's Mouse Models of Human Cancers Consortium. The authors also appreciate the discussions and review by Drs. A.D. Borowsky and J.A. Engelberg and support from Mr. Arishneel Ram.
- © 2014 Cold Spring Harbor Laboratory Press